brief
- seurat提供的教学里面包含了Standard pre-processing workflow,workflow包括QC,normalization,scale data ,detection of highly variable features。
- 其中 normalization就有蛮多方法的,seurat自己就提供了两种,一种是"LogNormalization",另一种是目前正在大力推荐的"SCTransform",而且每一种方法参数众多,我觉得有必要对其进行仔细的探究,所以需要分开学习和记录。
- 还有 detection of highly variable features以及scale data,也是方法和参数众多,我觉得有必要对其进行仔细的探究,所以需要分开学习和记录
- 概要图以及系列博文可以参见链接。
这里主要记录了 FindVariableFeatures的学习过程。
实例
library(dplyr)
library(Seurat)
library(patchwork)
library(sctransform)
rm(list=ls())
# 使用read10X读取output of the cellranger pipeline from 10X,包括barcodes,genes,matrix.mtx三个文件
pbmc.data <- Read10X(data.dir = "D:/djs/pbmc3k_filtered_gene_bc_matrices/filtered_gene_bc_matrices/hg19")
# 使用 CreateSeuratObject函数构造seurat对象
pbmc <- CreateSeuratObject(counts = pbmc.data, project = "pbmc3k",
min.cells = 3, min.features = 200,
names.delim = "-",names.field = 1)
pbmc[["percent.mt"]] <- PercentageFeatureSet(pbmc, pattern = "^MT-")
# Visualize QC metrics as a violin plot
VlnPlot(pbmc, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
# FeatureScatter is typically used to visualize feature-feature relationships, but can be used
# for anything calculated by the object, i.e. columns in object metadata, PC scores etc.
plot1 <- FeatureScatter(pbmc, feature1 = "nCount_RNA", feature2 = "percent.mt")
plot2 <- FeatureScatter(pbmc, feature1 = "nCount_RNA", feature2 = "nFeature_RNA")
plot1 + plot2
pbmc <- subset(pbmc, subset = nFeature_RNA > 200 & nFeature_RNA < 2500 & percent.mt < 5)
上面完成了Seurat对象的构建与QC.
==============================================================================================
下面开始进行normalization,然后开始 FindVariableFeatures
- normalization
pbmc <- NormalizeData(pbmc, normalization.method = "LogNormalize", scale.factor = 10000)
pbmc <- SCTransform(pbmc, vst.flavor = "v2", verbose = FALSE,variable.features.n = 2000)
str(pbmc)
# 此处我们直接把两种normalization方法都进运行了
# LogNormalize把数据放在了 assays$RNA@data
# SCTransform把数据放在了 assays$SCT@data,同时assays$SCT@counts与assays$RNA@counts也有区别
# SCTransform运行过程中已经做好了 FindVariableFeatures --> ScaleData ,分别放在了assays$SCT@scale.data /assays$SCT@var.features
- FindVariableFeatures
# 因为上面把两种normalization方法都进运行了,有两个data,所以需要指定assay = "RNA" / assay = "SCT"
pbmc <- FindVariableFeatures(pbmc,assay = "RNA" ,selection.method = "vst", nfeatures = 2000)
# Identification of highly variable features (feature selection)
# 这里得到的feature会在下面的pca降维种默认使用
# Identify the 10 most highly variable genes
head(VariableFeatures(pbmc,assay = "RNA"), 10)
head(VariableFeatures(pbmc,assay = "SCT"), 10)

这里你可以看到前十的高变基因就有差异了。
看到下面一个数据就更值得思考了。

- seurat提供的可视化方法
# plot variable features with and without labels
plot1 <- VariableFeaturePlot(pbmc,assay = "RNA")
plot2 <- LabelPoints(plot = plot1, points = top10, repel = TRUE)
plot1 + plot2
# plot variable features with and without labels
plot1 <- VariableFeaturePlot(pbmc,assay = "RNA")
plot2 <- VariableFeaturePlot(pbmc,assay = "SCT")
plot1 + plot2

然后variable 选择有几种方法,我试了一下不同方法在默认设置下的结果:
x <- intersect(VariableFeatures(pbmc_vst),VariableFeatures(pbmc_disp))
length(x)
length(VariableFeatures(pbmc_mvp))
x <- intersect(VariableFeatures(pbmc_vst),VariableFeatures(pbmc_mvp))
length(x)
x <- intersect(VariableFeatures(pbmc_disp),VariableFeatures(pbmc_mvp))
length(x)
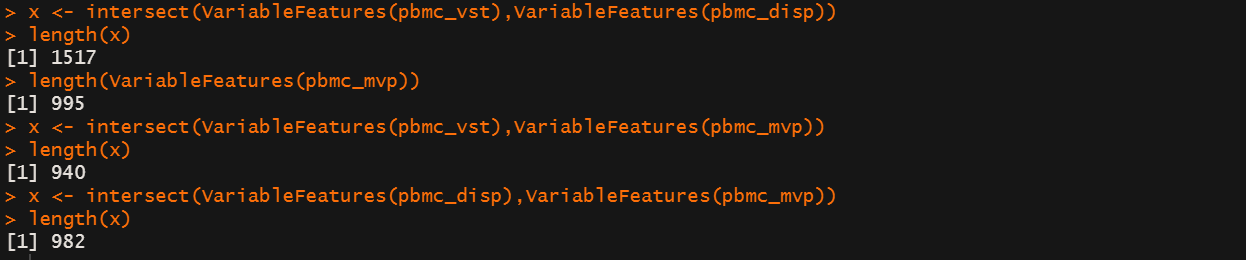

后续很多分析是基于variable features进行的,不同的normalization 流程和 feature selectiion对后续分析的具体影响暂时我还没搞清楚。
但是你可以看到normalization 流程的不同会影响 feature selectiion ⇒ feature selectiion 的方法不同会导致highly variable features不同 ⇒ highly variable features不同你的RunPCA and cluster以及annotation 都会有些区别。