目录
💥1 概述
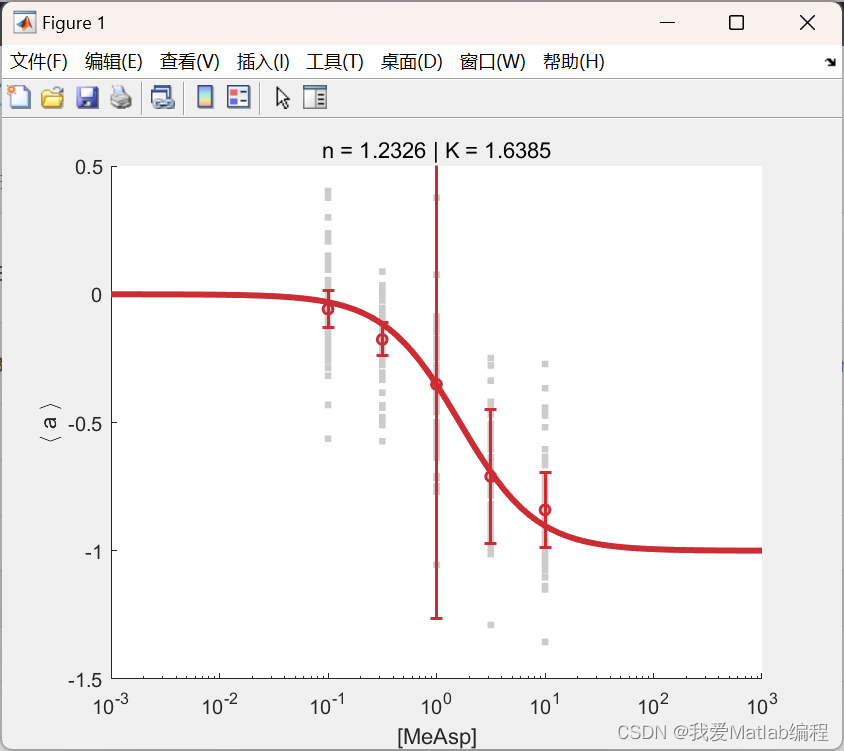
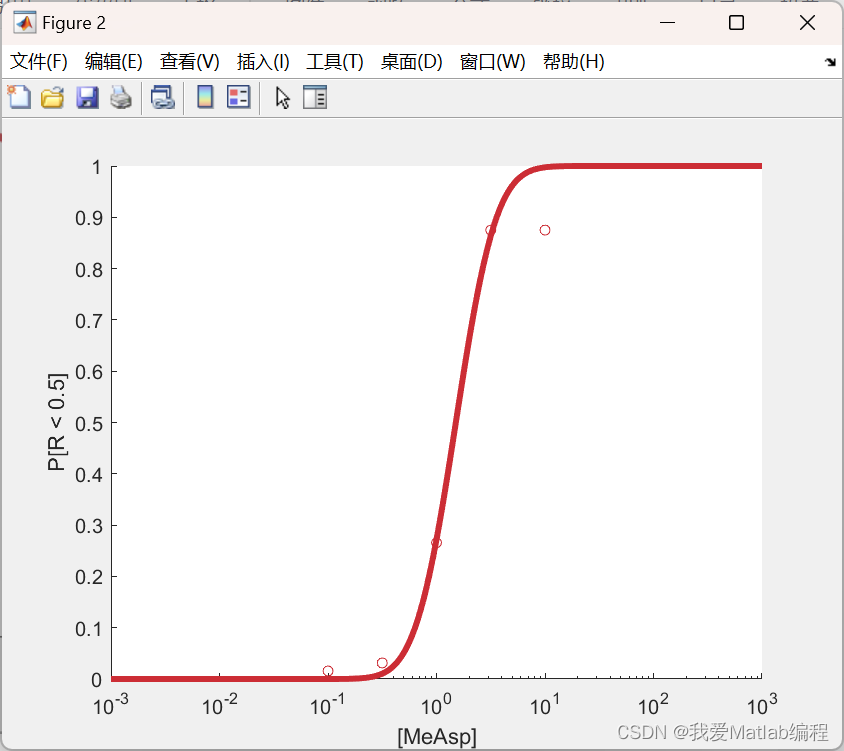
📚2 运行结果
🎉3 参考文献
👨💻4 Matlab代码
💥1 概述
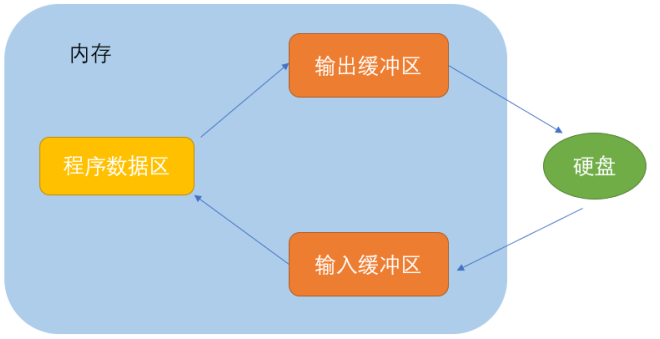
对于分子生物学来讲,生物分析手段的发展,是阐明机理的必要条件。在研究分子间相互作用的道路上,人们不断探索,总结出很多方法,免疫技术,晶体衍射,核磁共振等。1948年,荧光共振能量转移(Fluorescence resonance energy transfer,FRET)理论被首次提出,它可以测定1.0-6.0nm距离内分子间的相互作用。1967年,这一理论得到了实验验证,将1.0-6.0nm的距离称为光学尺。二十世纪八十年代出,通过科学家的不断探索,Fret技术成功运用到蛋白质结构的研究中。自Fret荧光共振能量技术诞生以来,已结合多种先进的技术和方法,如电子显微镜,X射线衍射等,推动了分子生物学检测手段的发展。
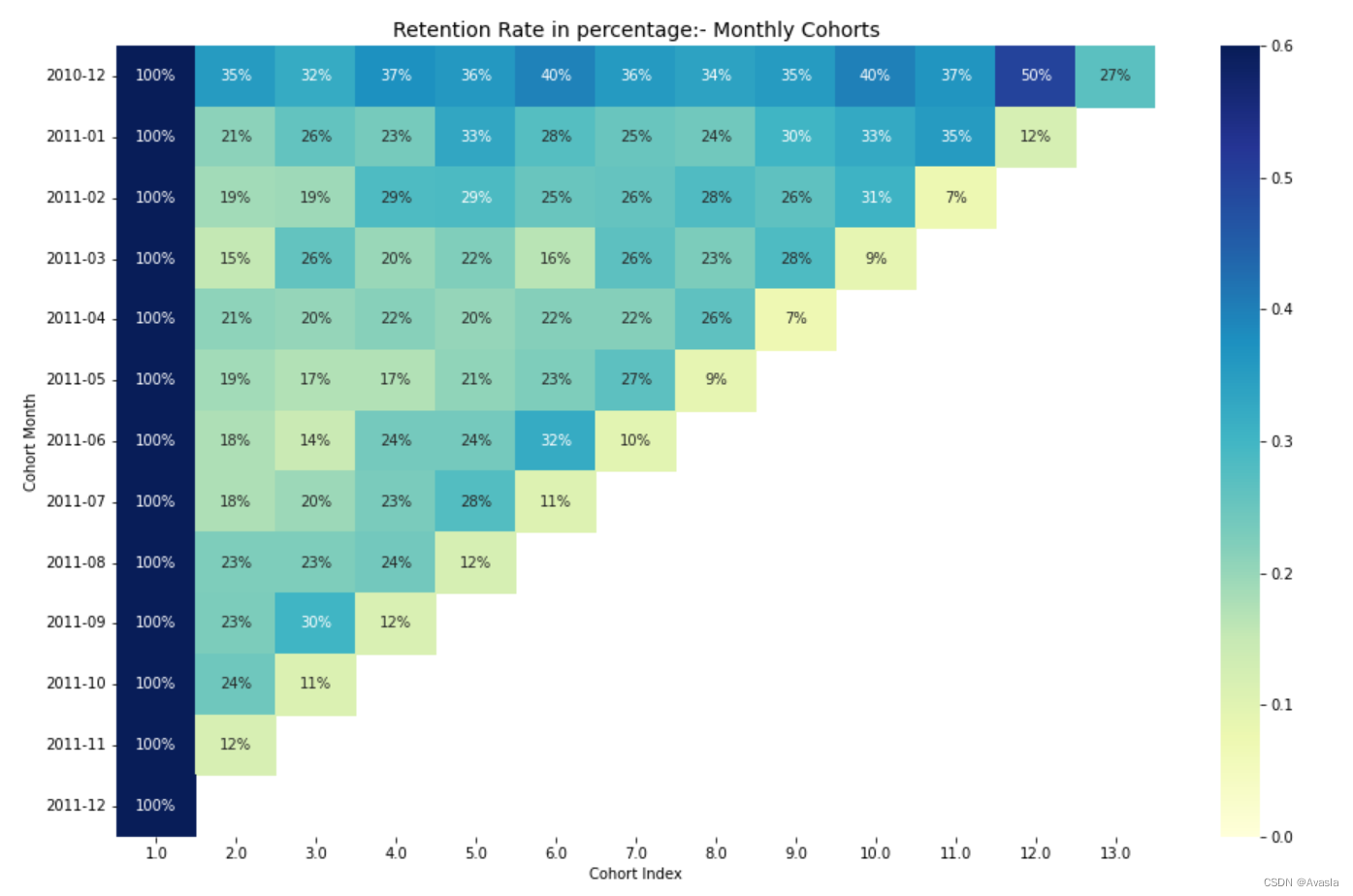
📚2 运行结果
🎉3 参考文献
[1]张建伟,陈同生.荧光共振能量转移(FRET)的定量检测及其应用[J].华南师范大学学报(自然科学版),2012,44(03):12-17.
👨💻4 Matlab代码
主函数部分代码:
%% Add necessary functions to the PATH
close all; clear all;
addpath('./Functions')
parms = parameters(); %Struct containing system parameters
showPlot = parms.showPlot;
%% Datasets: Load
dataparms = datasets(); %Load data parameters
backConc = dataparms.backConc;
xlabels = dataparms.xlabels;
Lplot = dataparms.Lplot;
Files = dataparms.Files;
OutFolder = dataparms.OutFolder;
concLevels = dataparms.concLevels;
%% Make output destination
OutputDest = ['./Outputs/', OutFolder];
mkdir(OutputDest)
%% Initial data processing/reformatting
EfretData = combineFretFiles(Files); %Combine files into one dataset
n = length(concLevels); %Number of stimulus levels
m = min(round(size(EfretData(1).A, 1)./n), parms.m); %Number of responses per stim level
%Reformat data into a struct with n matrices with m measurements each
doseData = reorganizeData(EfretData, n, m);
%Calculate response-amplitude for each individual stimulus
doseData = calcResponseAmp2(doseData, n, parms);
%Calculate population-average and single-cell dose-response data
[popAvgData, SCAvgData] = calcPopAvgDoseResponse2(doseData, parms);
%% Fit hill function to population-average dose-response data
%Collect dose-response data, and optimal parameters for hill function
HillPlotData = fitHillFunction(popAvgData, SCAvgData, dataparms, parms, OutputDest);
%% Fit CDF
CDFPlotData = fitLogNormCDF(SCAvgData, dataparms, parms, OutputDest);