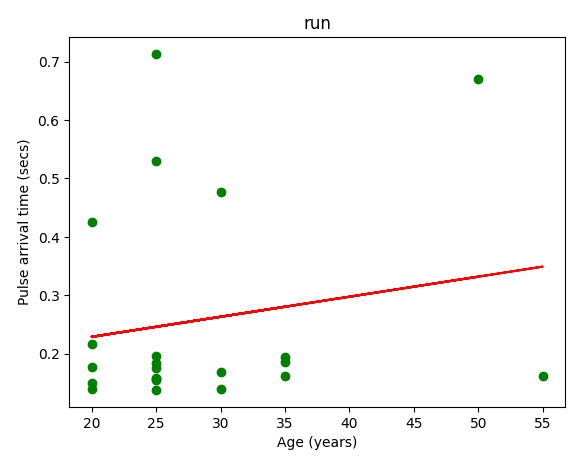
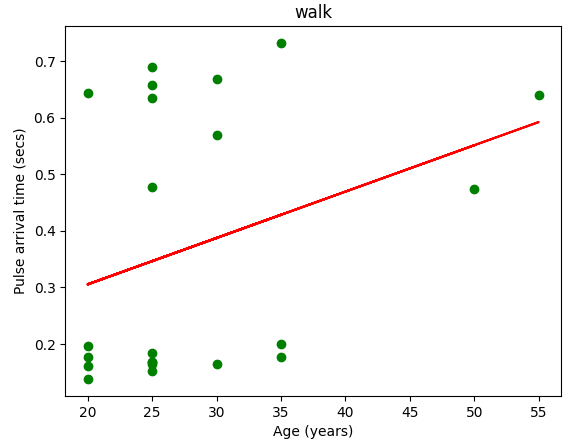
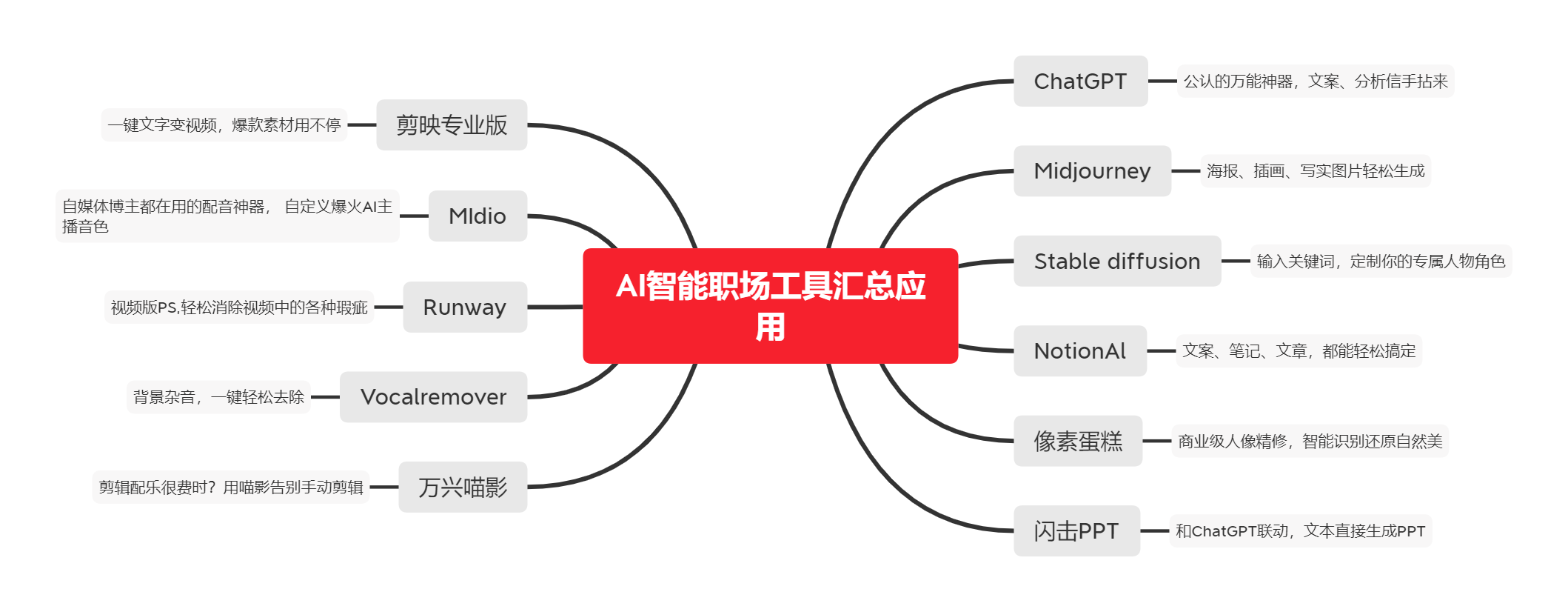
PAT 通常用作动脉硬度的间接测量值或心血管健康的指标。它与各种生理和病理状况有关,例如高血压、动脉硬化和内皮功能障碍。
通过脉搏到达时间进行测量,简单来说就是 先从脉冲传输时间 PPG 数据集中提取数据,提取此数据集中每个对象的脉冲到达时间,再提取参考年龄,进行一些比较。
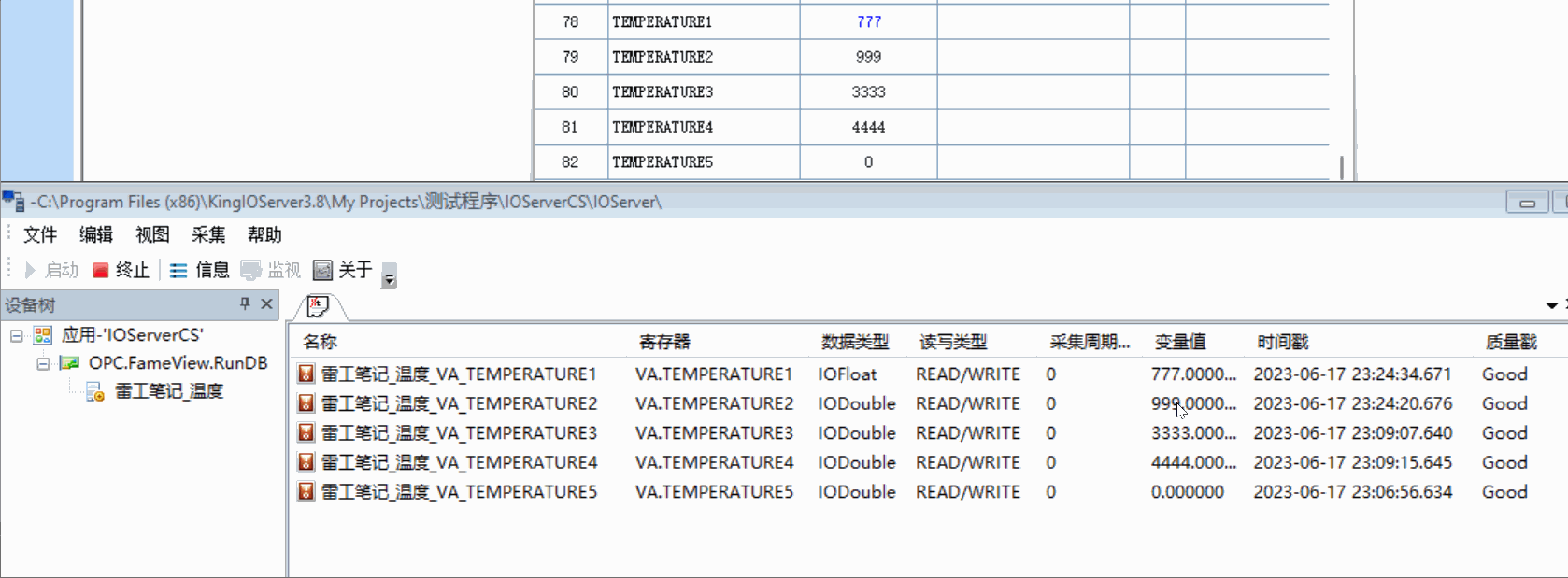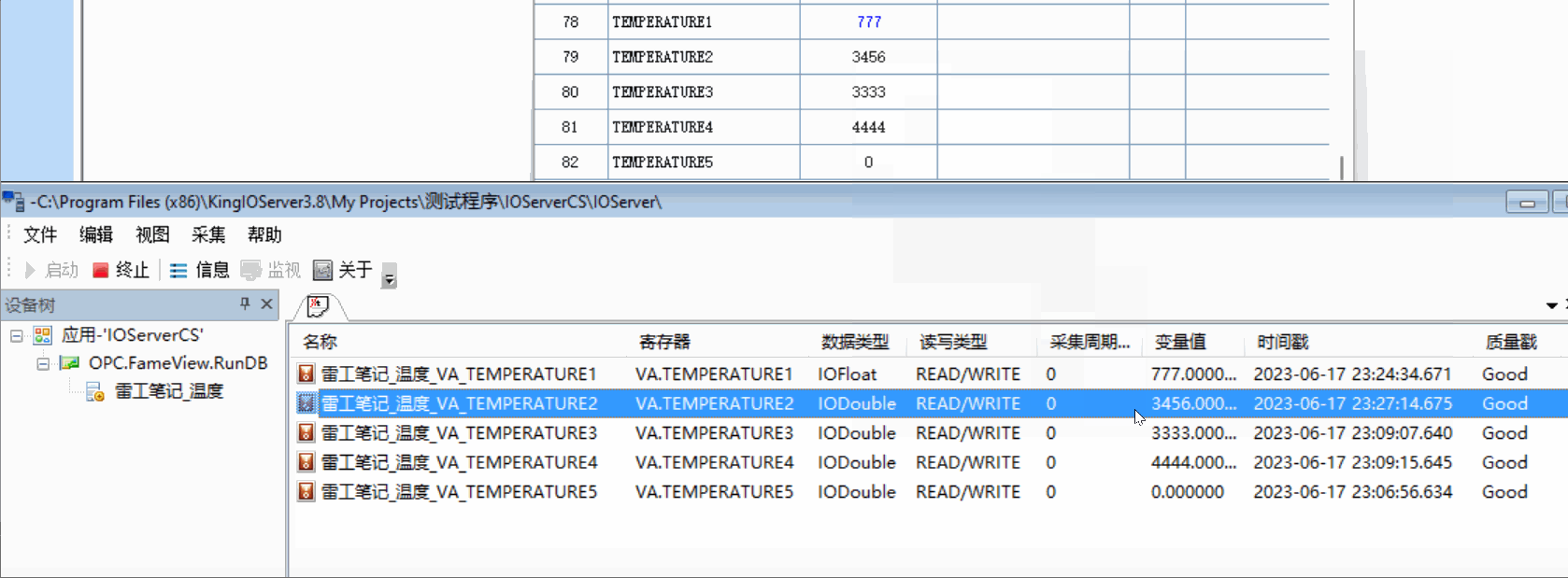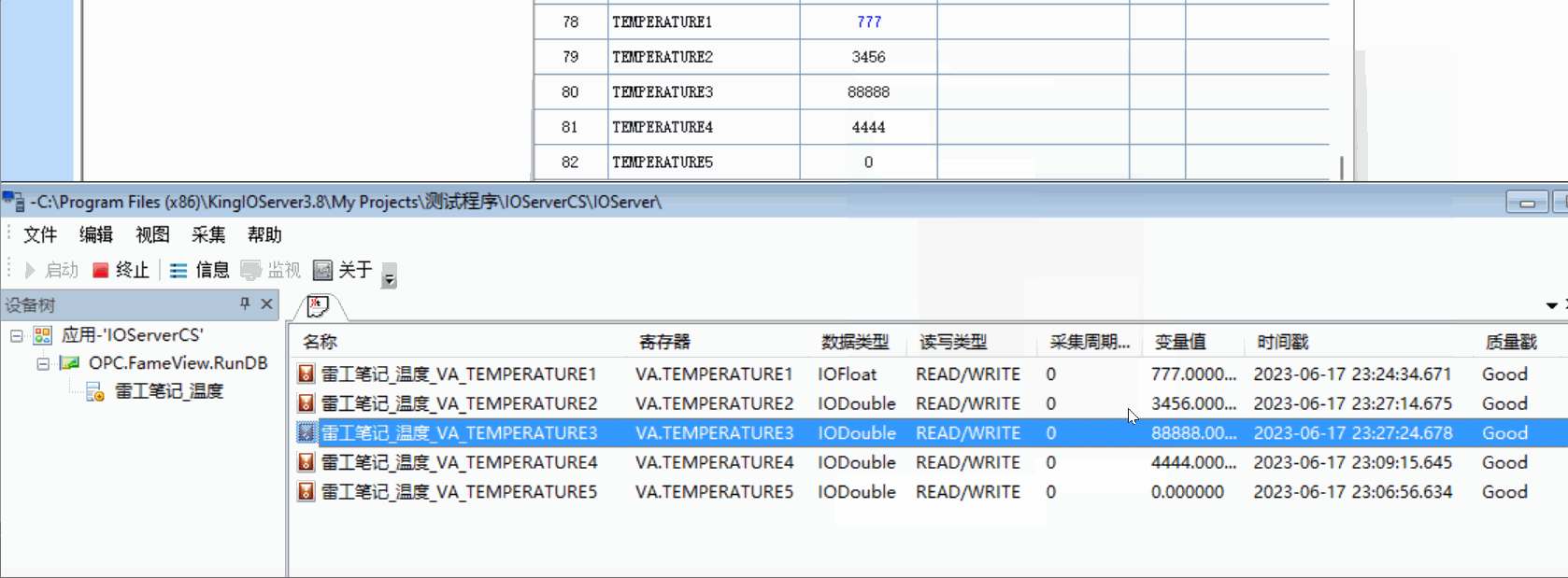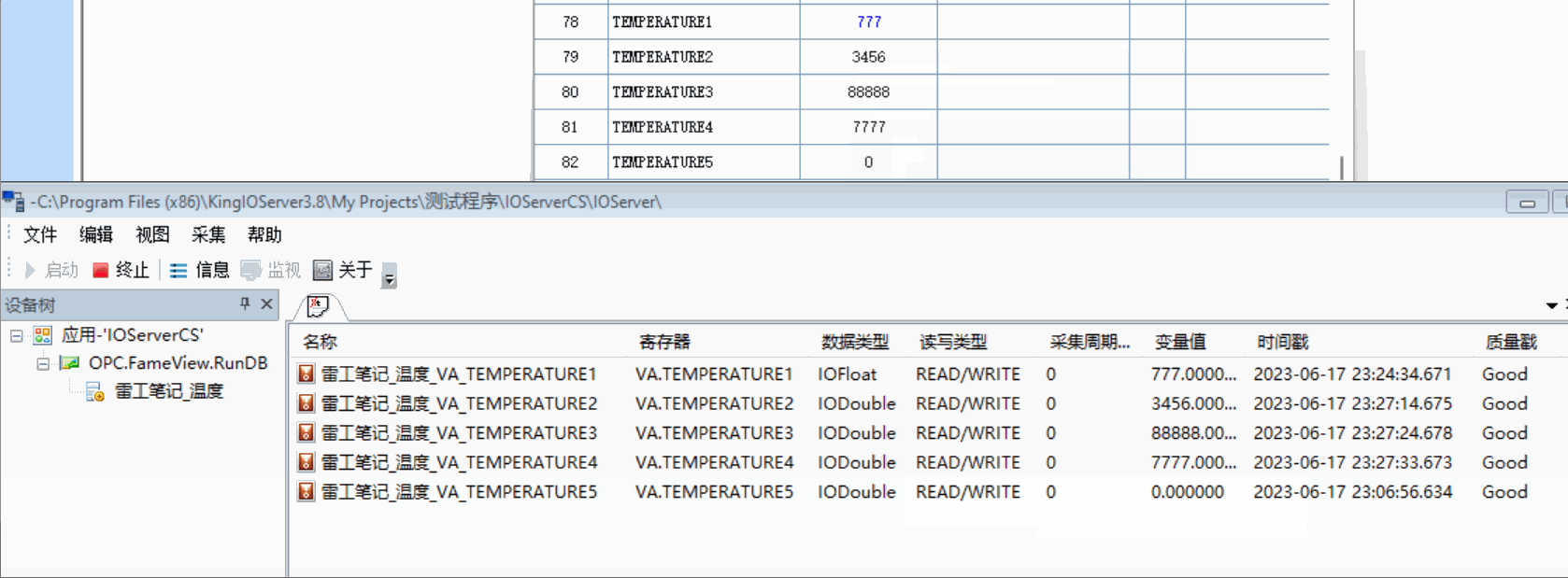
用的pulse transit time PPG数据集
22*3个数据记录了走跑坐
Bp_sys_start/end systolic blood pressure 收缩压
Bp_dia_start/end diastolic blood pressure 舒张压
Hr_1_start/end 测量开始时使用欧姆龙 HEM-7322 血压计测量的心率 (bpm)
Hr_2_start/end 测量开始时使用 iHealth Air 无线脉搏血氧仪测量的心率 (bpm)
spo2_start/end SpO2血氧饱和度
对于ecg和ppg信号,用neurokit2和wfdb库导入数据,
有一个需要注意的预处理操作是:
让ppg = -1*ppg,使ppg信号看起来更像血压信号。
再用neurokit2进行信号过滤,消除噪音
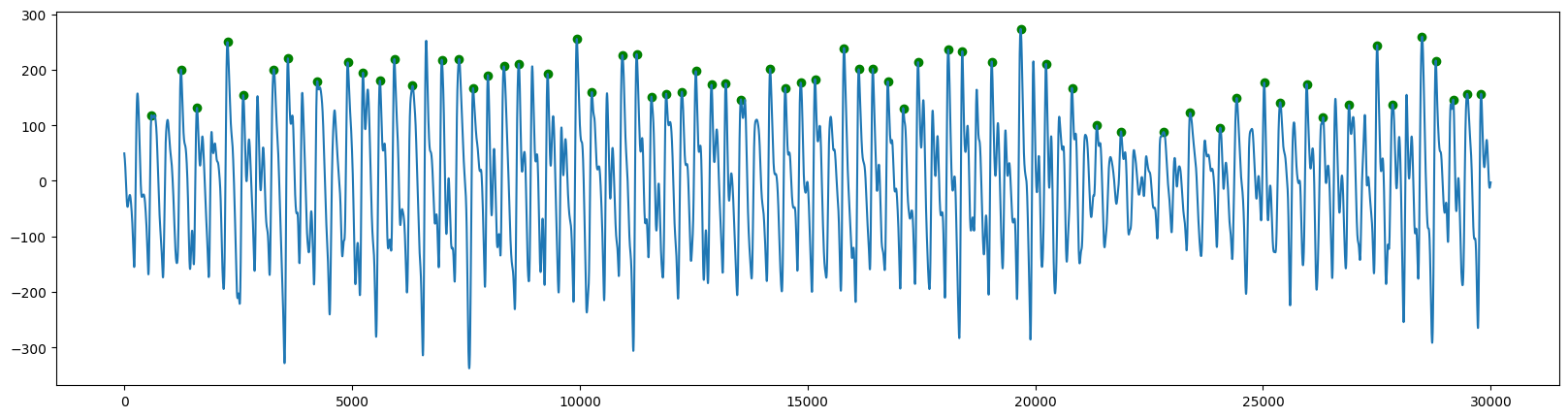
然后检测峰值:
ppg:
ecg:
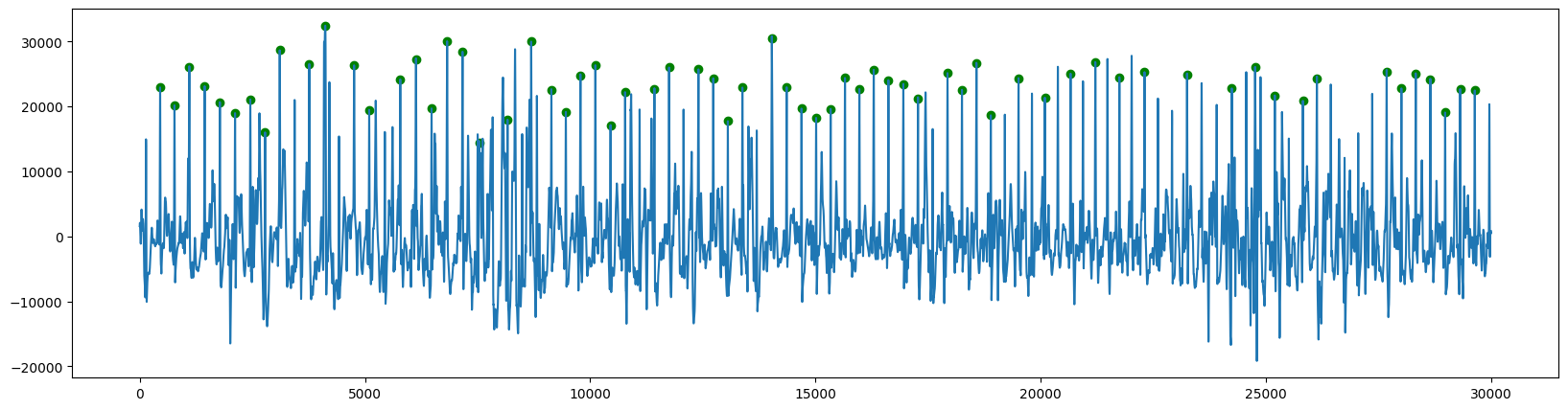
对于峰值时间的检测,ppg和ecg略有区别:
ecg可以直接提取R 波的时间,它表示心室去极化的时间(促使心室收缩的电活动)。
ppg则一般不使用收缩峰时间,因为脉搏开始和收缩峰之间可能存在可变延迟,所以一般会用脉冲开始时间
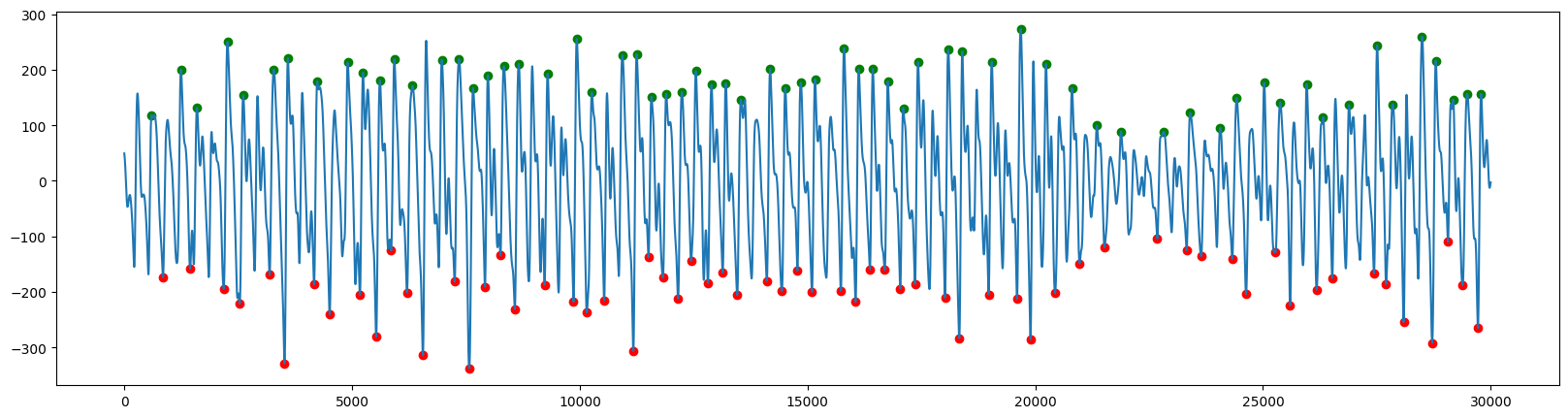
def get_ppg_onsets(ppg, pks, fs):
ons = np.empty(0)
for i in range(len(pks) - 1):
start = pks[i]
stop = pks[i + 1]
ibi = ppg[start:stop]
aux_ons = np.argmin(ibi)
ind_ons = aux_ons.astype(int)
ons = np.append(ons, ind_ons + start)
ons = ons.astype(int)
return ons
def get_data(record_list,database_name,required_signals,required_duration,required_activity):
matching_recs = {'dir': [], 'name': [], 'length': [], 'start_sbp': [], 'end_sbp': [], 'delta_sbp': [], 'age': []}
for record in record_list:
print('Record: {}'.format(record), end="", flush=True)
record_data = wfdb.rdheader(record, pn_dir=database_name, rd_segments=True)
# Check whether the required signals are present in the record
sigs_present = record_data.sig_name
if not all(x in sigs_present for x in required_signals):
print(' (missing signals)')
continue
if not required_activity in record:
print(' (not required activity)')
continue
seg_length = record_data.sig_len / (record_data.fs)
if seg_length < required_duration:
print(f' (too short at {seg_length / 60:.1f} mins)')
continue
# This record does meet the requirements, so extract information and data from it
# Information
matching_recs['dir'].append(database_name)
matching_recs['name'].append(record_data.record_name)
matching_recs['length'].append(seg_length)
# Blood pressure measurements
start_el = record_data.comments[0].index('<bp_sys_start>: ') + len('<bp_sys_start>: ')
end_el = record_data.comments[0].index('<bp_sys_end>') - 1
matching_recs['start_sbp'].append(int(record_data.comments[0][start_el:end_el]))
start_el = record_data.comments[0].index('<bp_sys_end>: ') + len('<bp_sys_end>: ')
end_el = record_data.comments[0].index('<bp_dia_start>') - 1
matching_recs['end_sbp'].append(int(record_data.comments[0][start_el:end_el]))
matching_recs['delta_sbp'].append(matching_recs['end_sbp'][-1] - matching_recs['start_sbp'][-1])
# ages
start_el = record_data.comments[0].index('<age>: ') + len('<age>: ')
end_el = record_data.comments[0].index('<bp_sys_start>') - 1
matching_recs['age'].append(float(record_data.comments[0][start_el:end_el]))
print(' (met requirements)')
print(f"A total of {len(matching_recs['dir'])} out of {len(record_list)} records met the requirements.")
return matching_recs
def get_pat(matching_recs,start_seconds,n_seconds_to_load,required_signals):
subj_nos = [i for i in range(len(matching_recs['name']))]
median_pats = []
for subj_no in subj_nos:
# specify this subject's record name and directory:
record_name = matching_recs['name'][subj_no]
record_dir = matching_recs['dir'][subj_no]
# extract this subject's signals
record_data = wfdb.rdheader(record_name, pn_dir=record_dir, rd_segments=True)
fs = record_data.fs
# Specify timings of segment to be extracted
sample_start = fs * start_seconds
sample_end = fs * (start_seconds + n_seconds_to_load)
# Load segment data
segment_data = wfdb.rdrecord(record_name=record_name,
channel_names=required_signals,
sampfrom=sample_start,
sampto=sample_end,
pn_dir=record_dir)
ppg_col = segment_data.sig_name.index("pleth_1")
ppg_final = segment_data.p_signal[:, ppg_col]
ecg_col = segment_data.sig_name.index("ecg")
ecg_final = segment_data.p_signal[:, ecg_col]
fs = segment_data.fs
ppg = -1 * ppg_final
# filter signals
ppg = nk2.ppg_clean(ppg, sampling_rate=fs)
ecg = nk2.ecg_clean(ecg_final, sampling_rate=fs)
# detect beats in signals
ppg_clean = nk2.ppg_clean(ppg, sampling_rate=fs)
ppg_peaks = nk2.ppg_findpeaks(ppg_clean, method="elgendi", show=False)['PPG_Peaks']
ecg_clean = nk2.ecg_clean(ecg, sampling_rate=fs)
ecg_signals, ecg_info = nk2.ecg_peaks(ecg_clean, method="neurokit", show=False)
ecg_peaks = ecg_info["ECG_R_Peaks"]
# obtain timings
ecg_timings = ecg_peaks
ppg_timings = get_ppg_onsets(ppg, ppg_peaks, fs)
# extract pulse arrival times
rel_ppg_timings = []
for ecg_timing in ecg_timings:
ppg_timings = np.asarray(ppg_timings)
different = ppg_timings - ecg_timing
different = np.where(different > 0, different, 100000)
idx = different.argmin()
rel_ppg_timings.append(ppg_timings[idx])
pats = (rel_ppg_timings - ecg_timings) / fs
# find median pulse arrival time for this subject
curr_median_pat = median(pats)
median_pats.append(curr_median_pat)
return median_pats
def get_age(matching_recs):
ages = []
subj_nos = [i for i in range(len(matching_recs['name']))]
for subj_no in subj_nos:
# specify this subject's record name and directory:
record_name = matching_recs['name'][subj_no]
record_dir = matching_recs['dir'][subj_no]
# extract this subject's age
curr_age = matching_recs['age'][subj_no]
ages.append(curr_age)
return ages
分别比较走,跑,坐