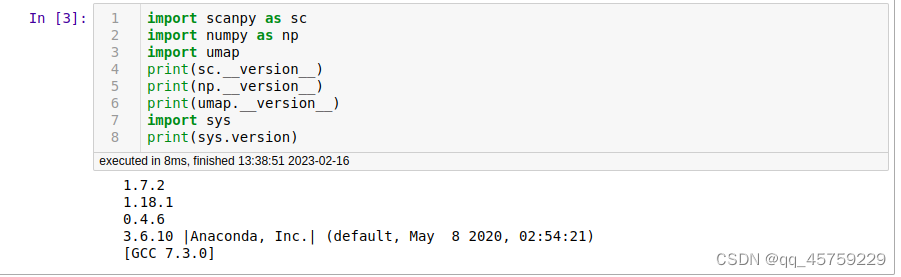
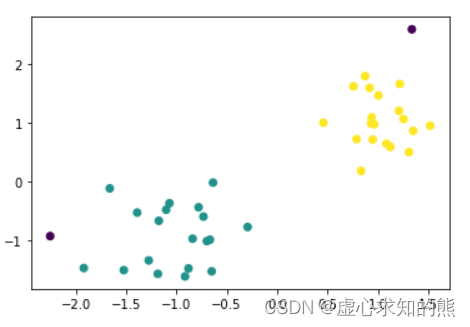
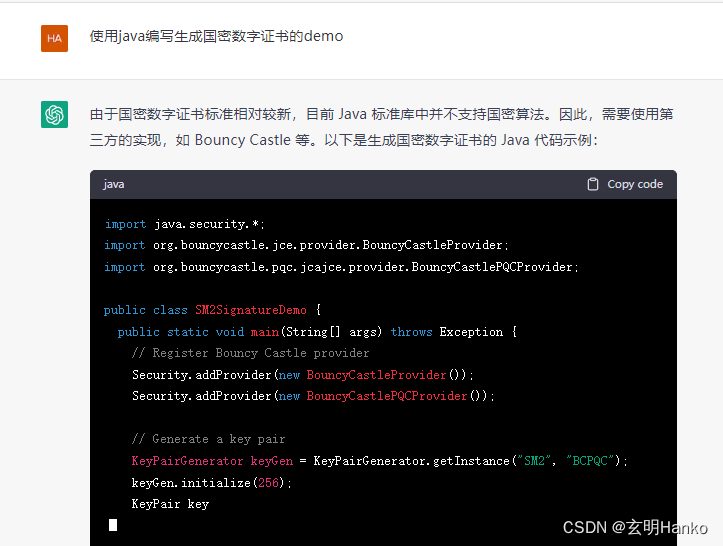
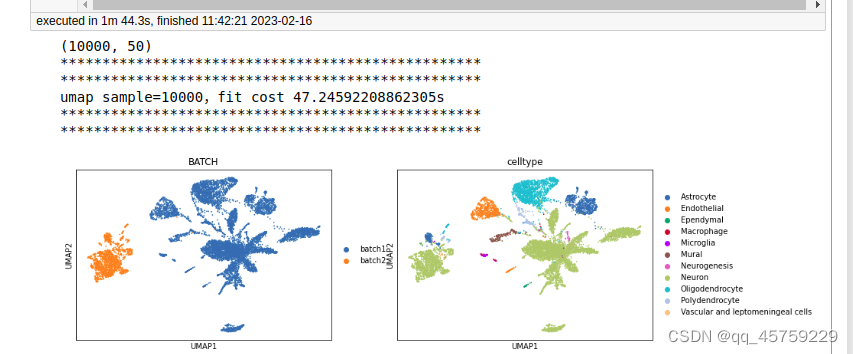
今天终于把umap的速度给测了一下,结果如下
预处理文件(83万的数据量)
# import scanpy as sc
# adata=sc.read("/DATA1/zhangjingxiao/yxk/dataset/FullMouseBrain/FullMouseBrain_raw.h5ad")
# print(adata)
# sc.pp.normalize_total(adata,target_sum=10000)
# sc.pp.log1p(adata)
# sc.pp.highly_variable_genes(adata, n_top_genes=2000, subset = True)
# sc.pp.scale(adata)
# sc.tl.pca(adata,svd_solver='arpack')
# #write to PCA h5ad
# adata.write("/DATA1/zhangjingxiao/yxk/dataset/FullMouseBrain/FullMouseBrain_pca.h5ad")
sample=10000
import scanpy as sc
#import hnswlib
import scanpy as sc
import pandas as pd
from time import time
# 怎么关闭warning
import os
os.environ['TF_CPP_MIN_LOG_LEVEL']='2'
############################
n_sample=10000
############################
adata=sc.read("/DATA1/zhangjingxiao/yxk/dataset/FullMouseBrain/FullMouseBrain_pca.h5ad")
adata=sc.pp.subsample(adata,n_obs=n_sample,copy=True)
print(adata.obsm["X_pca"].shape)
t0=time()
sc.pp.neighbors(adata)
sc.tl.umap(adata)
t1=time()
print("*"*50)
print("*"*50)
print("umap sample={},fit cost {}s".format(n_sample,t1-t0))
print("*"*50)
print("*"*50)
sc.pl.umap(adata,color = ["BATCH", "celltype"])
sample=20000
import scanpy as sc
#import hnswlib
import scanpy as sc
import pandas as pd
from time import time
# 怎么关闭warning
import os
os.environ['TF_CPP_MIN_LOG_LEVEL']='2'
############################
n_sample=20000
############################
adata=sc.read("/DATA1/zhangjingxiao/yxk/dataset/FullMouseBrain/FullMouseBrain_pca.h5ad")
adata=sc.pp.subsample(adata,n_obs=n_sample,copy=True)
print(adata.obsm["X_pca"].shape)
t0=time()
sc.pp.neighbors(adata)
sc.tl.umap(adata)
t1=time()
print("*"*50)
print("*"*50)
print("umap sample={},fit cost {}s".format(n_sample,t1-t0))
print("*"*50)
print("*"*50)
sc.pl.umap(adata,color = ["BATCH", "celltype"])

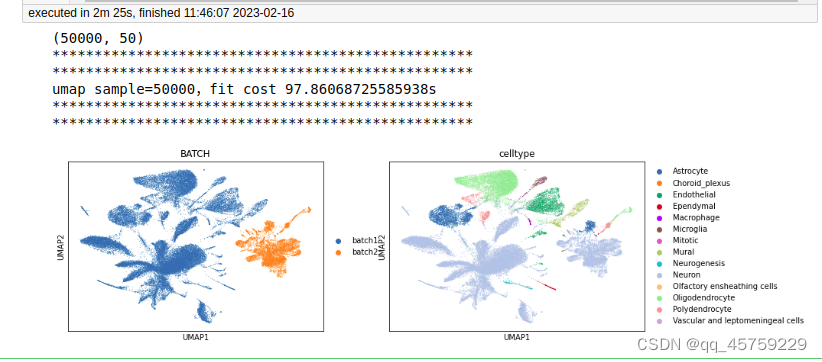
sample=50000
import scanpy as sc
#import hnswlib
import scanpy as sc
import pandas as pd
from time import time
# 怎么关闭warning
import os
os.environ['TF_CPP_MIN_LOG_LEVEL']='2'
############################
n_sample=50000
############################
adata=sc.read("/DATA1/zhangjingxiao/yxk/dataset/FullMouseBrain/FullMouseBrain_pca.h5ad")
adata=sc.pp.subsample(adata,n_obs=n_sample,copy=True)
print(adata.obsm["X_pca"].shape)
t0=time()
sc.pp.neighbors(adata)
sc.tl.umap(adata)
t1=time()
print("*"*50)
print("*"*50)
print("umap sample={},fit cost {}s".format(n_sample,t1-t0))
print("*"*50)
print("*"*50)
sc.pl.umap(adata,color = ["BATCH", "celltype"])
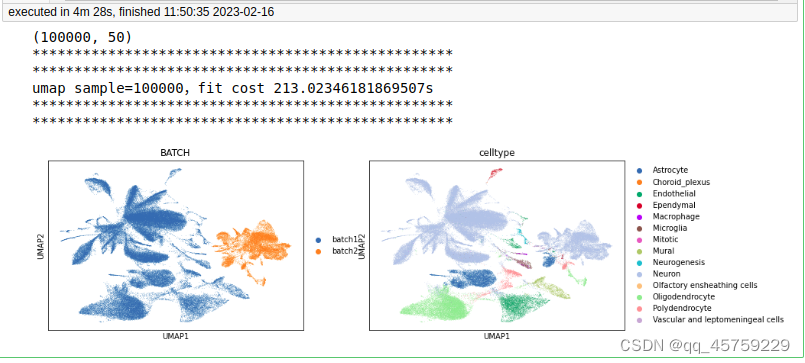
sample=100000
import scanpy as sc
#import hnswlib
import scanpy as sc
import pandas as pd
from time import time
# 怎么关闭warning
import os
os.environ['TF_CPP_MIN_LOG_LEVEL']='2'
############################
n_sample=100000
############################
adata=sc.read("/DATA1/zhangjingxiao/yxk/dataset/FullMouseBrain/FullMouseBrain_pca.h5ad")
adata=sc.pp.subsample(adata,n_obs=n_sample,copy=True)
print(adata.obsm["X_pca"].shape)
t0=time()
sc.pp.neighbors(adata)
sc.tl.umap(adata)
t1=time()
print("*"*50)
print("*"*50)
print("umap sample={},fit cost {}s".format(n_sample,t1-t0))
print("*"*50)
print("*"*50)
sc.pl.umap(adata,color = ["BATCH", "celltype"])
sample=200000
import scanpy as sc
#import hnswlib
import scanpy as sc
import pandas as pd
from time import time
# 怎么关闭warning
import os
os.environ['TF_CPP_MIN_LOG_LEVEL']='2'
############################
n_sample=200000
############################
adata=sc.read("/DATA1/zhangjingxiao/yxk/dataset/FullMouseBrain/FullMouseBrain_pca.h5ad")
adata=sc.pp.subsample(adata,n_obs=n_sample,copy=True)
print(adata.obsm["X_pca"].shape)
t0=time()
sc.pp.neighbors(adata)
sc.tl.umap(adata)
t1=time()
print("*"*50)
print("*"*50)
print("umap sample={},fit cost {}s".format(n_sample,t1-t0))
print("*"*50)
print("*"*50)
sc.pl.umap(adata,color = ["BATCH", "celltype"])

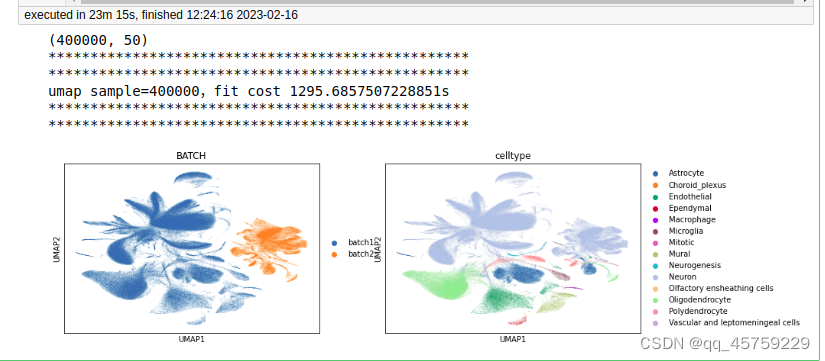
sample=400000
import scanpy as sc
#import hnswlib
import scanpy as sc
import pandas as pd
from time import time
# 怎么关闭warning
import os
os.environ['TF_CPP_MIN_LOG_LEVEL']='2'
############################
n_sample=400000
############################
adata=sc.read("/DATA1/zhangjingxiao/yxk/dataset/FullMouseBrain/FullMouseBrain_pca.h5ad")
adata=sc.pp.subsample(adata,n_obs=n_sample,copy=True)
print(adata.obsm["X_pca"].shape)
t0=time()
sc.pp.neighbors(adata)
sc.tl.umap(adata)
t1=time()
print("*"*50)
print("*"*50)
print("umap sample={},fit cost {}s".format(n_sample,t1-t0))
print("*"*50)
print("*"*50)
sc.pl.umap(adata,color = ["BATCH", "celltype"])
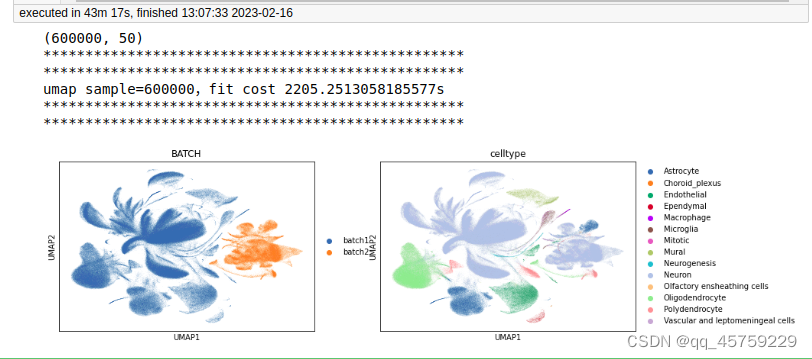
sample=600000
import scanpy as sc
#import hnswlib
import scanpy as sc
import pandas as pd
from time import time
# 怎么关闭warning
import os
os.environ['TF_CPP_MIN_LOG_LEVEL']='2'
############################
n_sample=600000
############################
adata=sc.read("/DATA1/zhangjingxiao/yxk/dataset/FullMouseBrain/FullMouseBrain_pca.h5ad")
adata=sc.pp.subsample(adata,n_obs=n_sample,copy=True)
print(adata.obsm["X_pca"].shape)
t0=time()
sc.pp.neighbors(adata)
sc.tl.umap(adata)
t1=time()
print("*"*50)
print("*"*50)
print("umap sample={},fit cost {}s".format(n_sample,t1-t0))
print("*"*50)
print("*"*50)
sc.pl.umap(adata,color = ["BATCH", "celltype"])
sample=800000
这个发生错误,因为计算sc.pp.neighbors()太耗内存了,内存直接炸掉了,所以出不来结果,不过这个时间应该在一个小时左右
环境如下