1. 颅骨去除
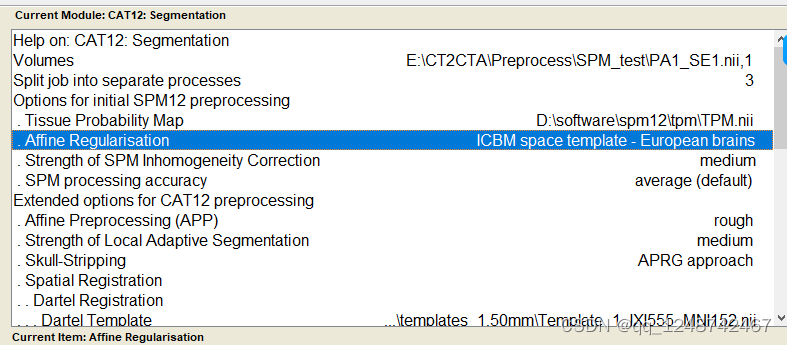

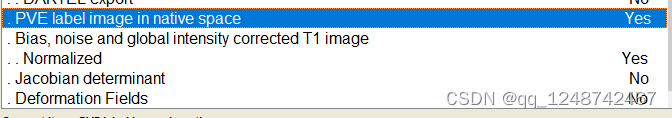
出现这个界面就一直等待即可:
segment的结果文件中会出现四个文件夹label、mri、report、surf
在mri文件中,mwp1是分割出来的灰质图像,mwp2是分割出来的白质图像,这两图像均是bias correction和空间配准后的。p0**应该是灰质+白质+脑脊液的图像,图像在原始空间上,没有配准。最后一个数据应该是去除头骨后的图像,bias correction和空间配准后的。
2. dicom和nifti互转
import dicom2nifti
import SimpleITK as itk
# pytorch311
# dicom2nifti.dicom_series_to_nifti("CTA-GAN/PA1/SE1", "CTA-GAN/PA1/SE1.nii.gz", reorient_nifti=True)
# nifti2dicom 就使用3D slicer进行操作即可
for i in range (1, 11):
dicom0_file = "PA" + str(i) + "/SE0"
nifti0_path = "PA" + str(i) + "_SE0.nii"
dicom2nifti.dicom_series_to_nifti(dicom0_file, nifti0_path, reorient_nifti=True)
dicom1_file = "PA"+str(i)+"/SE1"
nifti1_path = "PA"+str(i)+"_SE1.nii"
dicom2nifti.dicom_series_to_nifti(dicom1_file, nifti1_path, reorient_nifti=True)
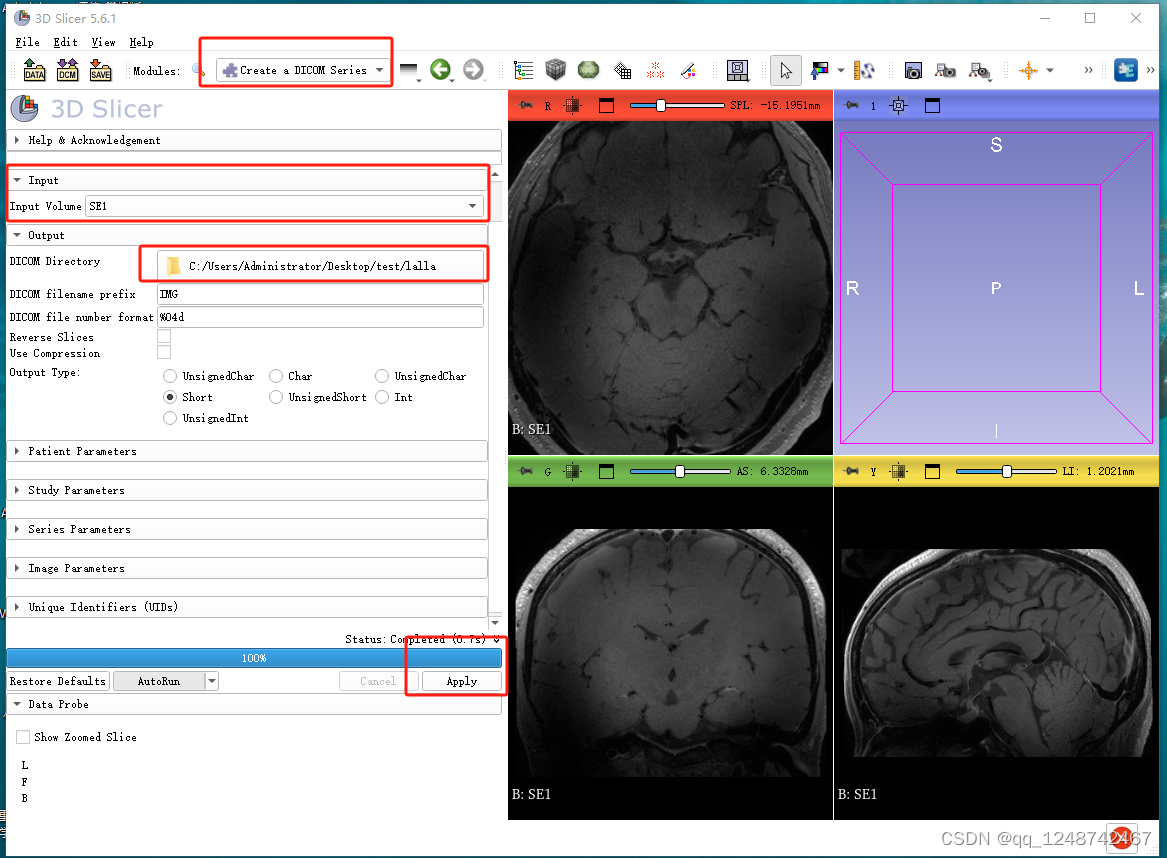
3. 配准到标准空间

Reference
- MRI使用SPM预处理