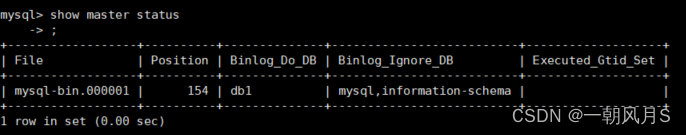
一、生成密度图
密度图标签生成
采用以下代码,生成训练集密度图gt:
import cv2
import glob
import h5py
import scipy
import pickle
import numpy as np
from PIL import Image
from itertools import islice
from tqdm import tqdm
from matplotlib import pyplot as plt
from sortedcontainers import SortedDict
from scipy.ndimage.filters import gaussian_filter
from scipy.spatial import KDTree
import argparse
"""
Code for DMnet, density map ground truth generation
Author: Changlin Li
Code revised on : 7/15/2020
Given dataset(train/val/test) generate ground truth for given dataset.
Default format for source data: The input images are in jpg format and raw annotations are in txt format
(Based on Visiondrone 2018/19/20 dataset)
Sample code to run:
python Generate_density_map_official.py . gaussian_kernels.pkl distances_dict.pkl --mode val
"""
# point_class_pair = {}
# annotation_stats = {0: 17, 1: 14, 2: 20, 3: 32, 4: 35, 5: 45, 6: 29, 7: 30, 8: 46, 9: 18}
# min_sigma, max_sigma = min(annotation_stats.values()), max(annotation_stats.values())
# print(min_sigma, max_sigma)
def get_img_paths(path_sets):
"""
Return all images from all pathes in 'path_sets'
"""
img_paths = []
for path in path_sets:
for img_path in glob.glob(os.path.join(path, '*.jpg')):
img_paths.append(img_path)
return img_paths
def save_computed_density(density_map, out_path):
"""
Save density map to h5py format
"""
with h5py.File(out_path, 'w') as hf:
hf['density'] = density_map
def compute_sigma(gt_count, distance=None, min_sigma=1, method=1, fixed_sigma=15):
"""
Compute sigma for gaussian kernel with different methods :
* method = 1 : sigma = (sum of distance to 3 nearest neighbors) / 10
* method = 2 : sigma = distance to nearest neighbor
* method = 3 : sigma = fixed value
** if sigma lower than threshold 'min_sigma', then 'min_sigma' will be used
** in case of one point on the image sigma = 'fixed_sigma'
"""
if gt_count > 1 and distance is not None:
if method == 1:
sigma = np.mean(distance[1:4]) * 0.1
elif method == 2:
sigma = distance[1]
elif method == 3:
sigma = fixed_sigma
else:
sigma = fixed_sigma
if sigma < min_sigma:
sigma = min_sigma
return sigma
def find_closest_key(sorted_dict, key):
"""
Find closest key in sorted_dict to 'key'
"""
keys = list(islice(sorted_dict.irange(minimum=key), 1))
keys.extend(islice(sorted_dict.irange(maximum=key, reverse=True), 1))
return min(keys, key=lambda k: abs(key - k))
def gaussian_filter_density(non_zero_points, map_h, map_w, distances=None, kernels_dict=None, min_sigma=2, method=1,
const_sigma=15):
"""
Fast gaussian filter implementation : using precomputed distances and kernels
"""
gt_count = non_zero_points.shape[0]
density_map = np.zeros((map_h, map_w), dtype=np.float32)
for i in range(gt_count):
point_x, point_y, category = non_zero_points[i]
sigma = compute_sigma(gt_count, distances[i], min_sigma=min_sigma, method=method, fixed_sigma=const_sigma)
# closest_sigma = annotation_stats[category]
closest_sigma = find_closest_key(kernels_dict, sigma)
# print(i,closest_sigma)
kernel = kernels_dict[closest_sigma]
full_kernel_size = kernel.shape[0]
kernel_size = full_kernel_size // 2
min_img_x = max(0, point_x - kernel_size)
min_img_y = max(0, point_y - kernel_size)
max_img_x = min(point_x + kernel_size + 1, map_w - 1)
max_img_y = min(point_y + kernel_size + 1, map_h - 1)
assert max_img_x > min_img_x
assert max_img_y > min_img_y
kernel_x_min = kernel_size - point_x if point_x <= kernel_size else 0
kernel_y_min = kernel_size - point_y if point_y <= kernel_size else 0
kernel_x_max = kernel_x_min + max_img_x - min_img_x
kernel_y_max = kernel_y_min + max_img_y - min_img_y
assert kernel_x_max > kernel_x_min
assert kernel_y_max > kernel_y_min
density_map[min_img_y:max_img_y, min_img_x:max_img_x] += kernel[kernel_y_min:kernel_y_max,
kernel_x_min:kernel_x_max]
return density_map
def get_gt_dots(ann_path, img_height, img_width, mode="train"):
"""
Load Matlab file with ground truth labels and save it to numpy array.
** cliping is needed to prevent going out of the array
"""
txt_list = open(ann_path, 'r').readlines()
gt = format_label(mode, txt_list)
assert gt.shape[1] == 3
gt[:, 0] = gt[:, 0].clip(0, img_width - 1)
gt[:, 1] = gt[:, 1].clip(0, img_height - 1)
return gt
def set_circles_on_img(image, bbox_list, circle_size=2):
"""
Set circles on images at centers of bboxes in bbox_list
"""
for bbox in bbox_list:
cv2.circle(image, (bbox[0], bbox[1]), circle_size, (255, 0, 0), -1)
return image
def generate_gaussian_kernels(out_kernels_path='gaussian_kernels.pkl', round_decimals=3, sigma_threshold=4, sigma_min=0,
sigma_max=20, num_sigmas=801):
"""
Computing gaussian filter kernel for sigmas in linspace(sigma_min, sigma_max, num_sigmas) and saving
them to dict.
"""
if os.path.exists(out_kernels_path):
# If kernel has been pre-computed, then return
print("Kernel already created!\nExiting...\n")
return
kernels_dict = dict()
sigma_space = np.linspace(sigma_min, sigma_max, num_sigmas)
for sigma in tqdm(sigma_space):
sigma = np.round(sigma, decimals=round_decimals)
kernel_size = np.ceil(sigma * sigma_threshold).astype(np.int)
img_shape = (kernel_size * 2 + 1, kernel_size * 2 + 1)
img_center = (img_shape[0] // 2, img_shape[1] // 2)
arr = np.zeros(img_shape)
arr[img_center] = 1
arr = scipy.ndimage.filters.gaussian_filter(arr, sigma, mode='constant')
kernel = arr / arr.sum()
kernels_dict[sigma] = kernel
print(f'Computed {len(sigma_space)} gaussian kernels. Saving them to {out_kernels_path}')
with open(out_kernels_path, 'wb') as f:
pickle.dump(kernels_dict, f)
def compute_distances(out_dist_path='distances_dict.pkl', raw_label_dir='D:/BaiduNetdiskDownload/VisDrone', n_neighbors=4,
leafsize=1024, data_limit=None, mode="train", img_affix=".jpg"):
if os.path.exists(out_dist_path):
# If distance has been computed, then directly load distance file.
print("Distrance pre-computation already created!\nExiting...\n")
return
distances_dict = dict()
full_img_paths = glob.glob(f'{raw_label_dir}/VisDrone2019-DET-train/images/*' + img_affix) + \
glob.glob(f'{raw_label_dir}/VisDrone2019-DET-val/images/*' + img_affix) + \
glob.glob(f'{raw_label_dir}/VisDrone2019-DET-test-dev/images/*' + img_affix)
if data_limit and data_limit < len(full_img_paths):
full_img_paths = full_img_paths[:data_limit]
for img_path in tqdm(full_img_paths):
ann_path = img_path.replace(img_affix, '.txt')
ann_path = ann_path.replace("images", "annotations")
img = plt.imread(img_path)
non_zero_points = get_gt_dots(ann_path, *img.shape[0:2], mode=mode)
tree = KDTree(non_zero_points.copy(), leafsize=leafsize) # build kdtree
distances, _ = tree.query(non_zero_points, k=n_neighbors) # query kdtree
distances_dict[img_path] = distances
print(f'Distances computed for {len(full_img_paths)}. Saving them to {out_dist_path}')
with open(out_dist_path, 'wb') as f:
pickle.dump(distances_dict, f)
def format_label(mode, txt_list):
format_data = []
# required format: xmin, ymin, xmax, ymax, class_id, clockwise direction
# Given format: <bbox_left>,<bbox_top>,<bbox_width>,<bbox_height>,class_id
for idx, i in enumerate(txt_list):
coord_raw = [int(x) for x in i.replace("\n", "").split(',') if len(x) != 0]
coord = coord_raw[:6]
# print(coord)
if len(coord) != 6:
# 4 coord + 1 class
print("Failed to parse annotation!")
exit()
# if coord[-1] not in class_list and coord[-1]>len(class_list):
# print('warning found a new label :', coord[-1])
# exit()
if coord[2] <= 0 or coord[3] <= 0:
print("Error encountered!\nFind out 0 height(width)!")
print("This bounding box has been discarded! ")
continue
# print("Pull out corrd matrix:\n")
# print(coord)
# exit(-1)
if not 0 < coord[-1] < 11:
# class 0 and 11 are not in our interest
continue
if mode == "VisDrone2019-DET-val" or "VisDrone2019-DET-test":
# in this case, score is the last 2 element.
# No consideration for score 0 in eval
if int(coord[-2]) == 0:
continue
if int(coord_raw[-2]) == 2:
continue
bbox_left, bbox_top = coord[0], coord[1]
bbox_right, bbox_bottom = coord[0] + coord[2], coord[1] + coord[3]
# Scale class number back to range 0-9
center_x, center_y = int((bbox_left + bbox_right) * 0.5), int((bbox_top + bbox_bottom) * 0.5)
format_data.append([center_x, center_y, coord[-1] - 1])
# if not filename:
# continue
# if filename not in point_class_pair:
# point_class_pair[filename] = {}
# coord_pair = str(center_x) + " " + str(center_y)
# if coord_pair not in point_class_pair[filename]:
# point_class_pair[filename][coord_pair] = coord[-1] - 1
# else:
# if point_class_pair[filename][coord_pair] != coord[-1] - 1:
# assert True, \
# "duplicate coordination shows in current file : " + str(filename)
return np.array(format_data)
def parse_args():
parser = argparse.ArgumentParser(
description='DMNet--Density map ground truth generation')
parser.add_argument('root_dir', default=".",
help='the path for source data')
parser.add_argument('precomputed_kernels_path', default="gaussian_kernels.pkl",
help='the path to save precomputed kernels')
parser.add_argument('precomputed_distances_path', default="distances_dict.pkl",
help='the path to save precomputed distance')
parser.add_argument('--image_prefix', default=".jpg", help='the path to save precomputed distance')
parser.add_argument('--mode', default="train", help='Indicate if you are working on train/val/test set')
parser.add_argument('--showden', action='store_true', help='show results')
args = parser.parse_args()
return args
if __name__ == "__main__":
# General setup
args = parse_args()
data_limit = None
precomputed_kernels_path = args.precomputed_kernels_path
precomputed_distances_path = args.precomputed_distances_path
img_affix = args.image_prefix
showden = args.showden
mode = args.mode
root_dir = args.root_dir
min_sigma = 0
max_sigma = 20
# create dir to save train/val density map
if not os.path.exists(os.path.join('/root/autodl-tmp/VisDrone2019/VisDrone2019-DET-train', 'dens')):
os.makedirs(os.path.join('/root/autodl-tmp/VisDrone2019/VisDrone2019-DET-train', 'dens'), exist_ok=True)
if not os.path.exists(os.path.join('/root/autodl-tmp/VisDrone2019/VisDrone2019-DET-val', 'dens')):
os.makedirs(os.path.join('/root/autodl-tmp/VisDrone2019/VisDrone2019-DET-val', 'dens'), exist_ok=True)
if not os.path.exists(os.path.join('/root/autodl-tmp/VisDrone2019/VisDrone2019-DET-test-dev', 'dens')):
os.makedirs(os.path.join('/root/autodl-tmp/VisDrone2019/VisDrone2019-DET-test-dev', 'dens'), exist_ok=True)
# create pre-computed kernel to speed up density map generation
generate_gaussian_kernels(precomputed_kernels_path, round_decimals=3, sigma_threshold=4,
sigma_min=min_sigma, sigma_max=max_sigma, num_sigmas=801)
with open(precomputed_kernels_path, 'rb') as f:
kernels_dict = pickle.load(f)
kernels_dict = SortedDict(kernels_dict)
# uncomment to generate and save dict with distances
compute_distances(out_dist_path=precomputed_distances_path, raw_label_dir=root_dir, mode=mode)
with open(precomputed_distances_path, 'rb') as f:
distances_dict = pickle.load(f)
# print(distances_dict)
data_root = mode
img_paths = glob.glob(f'{root_dir}/{data_root}/images/*.jpg')
method = 3
const_sigma = 15
with open(str(mode) + ".txt", "w") as fileloader:
# Prepared for the training algorithms that requires a txt output file
# with all input images listed
for img_path in tqdm(img_paths):
fileloader.write(img_path)
fileloader.write("\n")
data_folder, img_sub_path = img_path.split('images')
ann_path = img_path.replace(img_affix, '.txt')
ann_path = ann_path.replace("images", 'annotations')
# load img and gt
img = Image.open(img_path)
# print(img_path)
width, height = img.size
gt_points = get_gt_dots(ann_path, height, width, mode=mode)
distances = distances_dict[img_path]
density_map = gaussian_filter_density(gt_points, height, width, distances,
kernels_dict, min_sigma=min_sigma, method=method,
const_sigma=const_sigma)
den_name = os.path.join(root_dir, data_root, "dens", img_path.split("/")[-1].replace("jpg", "npy"))
print(den_name)
if showden:
plt.imshow(img)
plt.imshow(density_map, alpha=0.75)
plt.show()
else:
np.save(den_name, density_map)
使用以下命令进行创建:
# 生成训练集gt密度图
python image_cropping/Generate_density_map_official.py /root/autodl-tmp/VisDrone2019 gaussian_kernels.pkl distances_dict.pkl --mode VisDrone2019-DET-train
# 生成验证集gt密度图
python image_cropping/Generate_density_map_official.py /root/autodl-tmp/VisDrone2019 gaussian_kernels.pkl distances_dict.pkl --mode VisDrone2019-DET-val
# 生成测试集gt密度图
python image_cropping/Generate_density_map_official.py /root/autodl-tmp/VisDrone2019 gaussian_kernels.pkl distances_dict.pkl --mode VisDrone2019-DET-test-dev
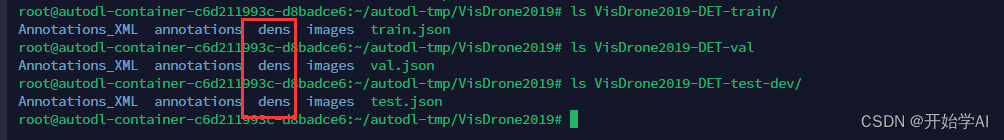
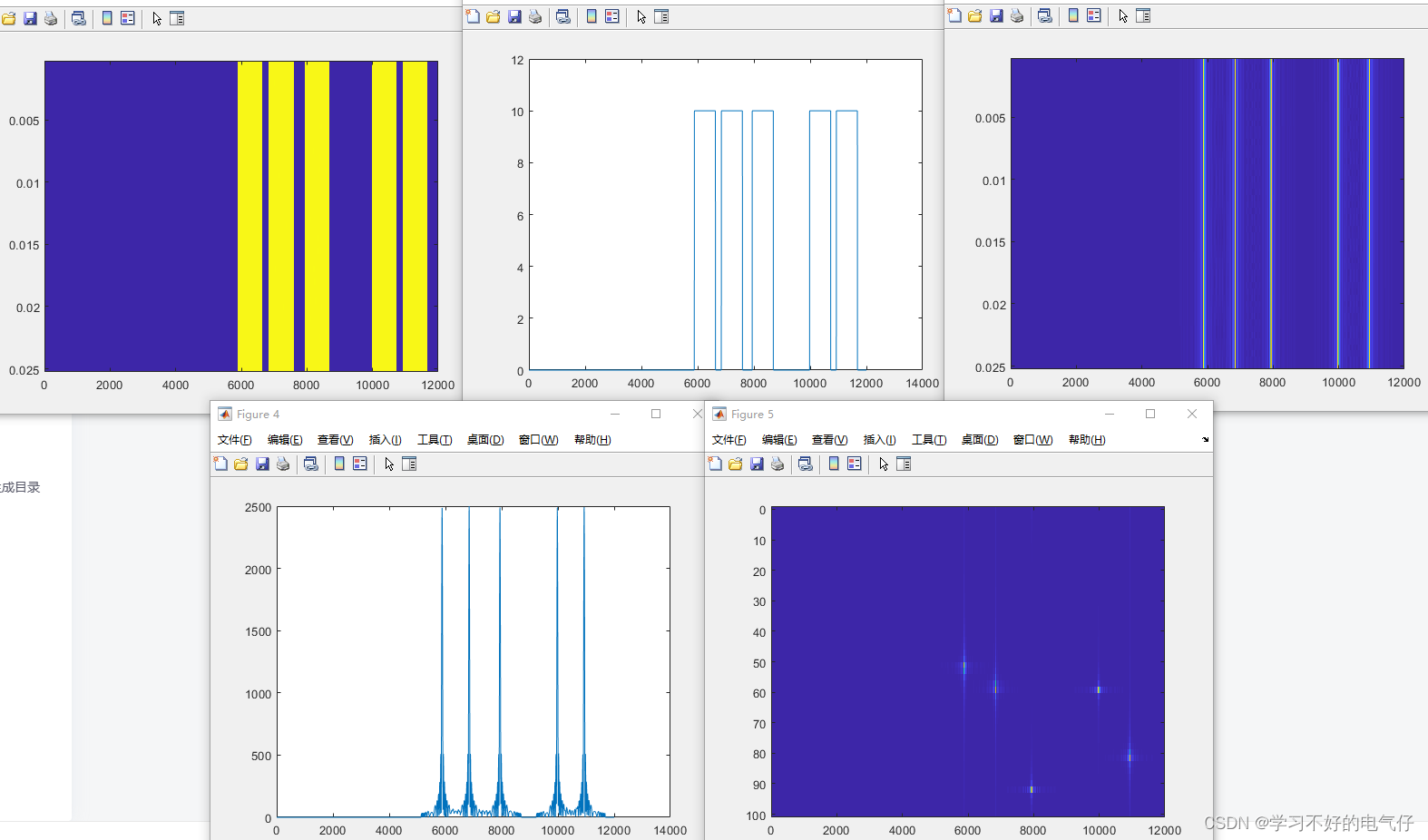
在以上文件夹 生成了如下的dens文件夹 保存了密度图的npy文件。
密度图预测
根据官方仓库说明 下载MCNN代码 进行训练预测即可
裁剪区域
# 生成训练集
python image_cropping/density_slide_window_official.py /root/autodl-tmp/VisDrone2019 70_70 0.08 --output_folder /root/autodl-tmp/VisDrone2019/Crop_70_0.08 --mode VisDrone2019-DET-train
# 生成验证集
python image_cropping/density_slide_window_official.py /root/autodl-tmp/VisDrone2019 70_70 0.08 --output_folder /root/autodl-tmp/VisDrone2019/Crop_70_0.08 --mode VisDrone2019-DET-val
# 生成测试集
python image_cropping/density_slide_window_official.py /root/autodl-tmp/VisDrone2019 70_70 0.08 --output_folder /root/autodl-tmp/VisDrone2019/Crop_70_0.08 --mode VisDrone2019-DET-test-dev
部分切分后的效果图,可以看到部分目标还是存在截断现象,是一个优化点。





由于不清楚是否会将crop后的图像与标签,用于到训练过程中,这里我就不生成训练的crop图和标签了,下一步准备直接用训练好的模型进行融合测试。论文是采用验证集进行测试的,这儿我准备把验证集和测试集(test-dev)都测试一遍。

最后按以下结构需要组织一下文件

最后我的目录结构如下
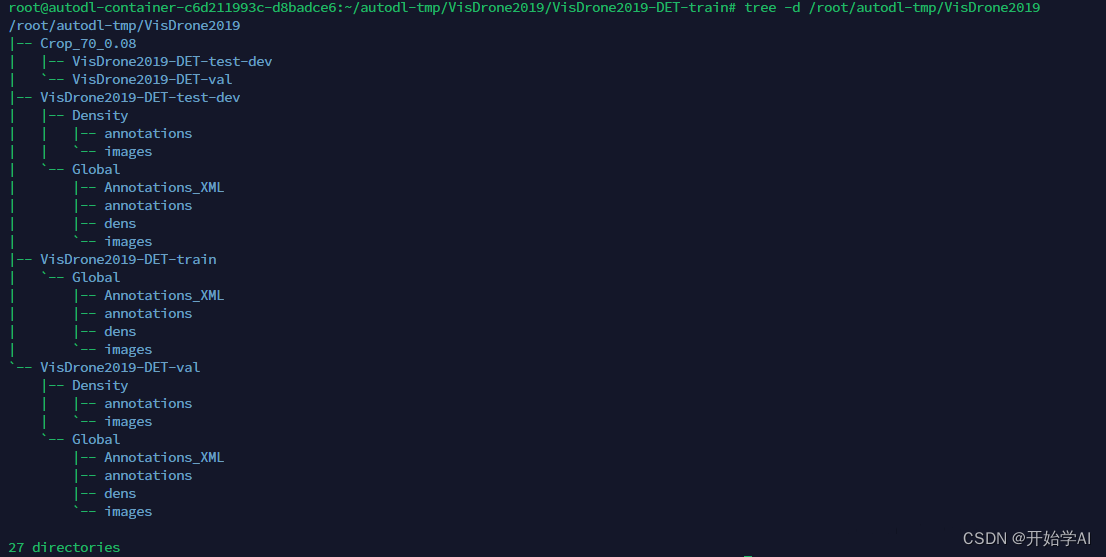
数据格式转换
需要先把txt转为voc,再把voc转为coco
# 生成测试集的voc格式
python fusion_detection/create_VOC_annotation_official.py /root/autodl-tmp/VisDrone2019 --output_folder VisDrone2019_finally --mode VisDrone2019-DET-test-dev
# 生成测试集的coco格式
python fusion_detection/VOC2coco_official.py /root/autodl-tmp/VisDrone2019/VisDrone2019_finally --mode VisDrone2019-DET-test-dev
这过程太耗时间了。最后把中间文件夹删除,整个格式如以下:
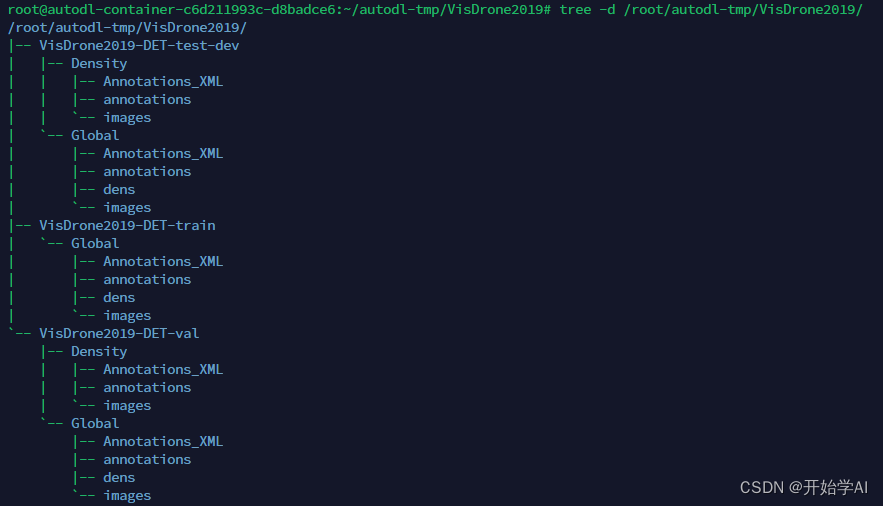
其中coco的json文件在以下位置
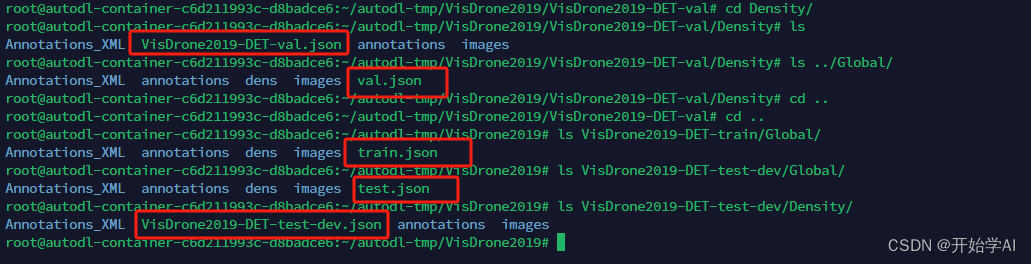
过程太繁琐了。
有需要生成后的数据,可以联系!