之前在博客中提到,会考虑用深度学习来对3D点云进行处理,接下来迈出脚步,先整几个例子来熟悉它。例子原型来源于官网,博主在其基础上做了一些代码修改。
一. 例子参考
1. Keras中的资源
Code examples

2.openvinotoolkit
open_model_zoo/demos at master · openvinotoolkit/open_model_zoo · GitHub
二. 例子实现
1.pointNet(Keras实现)
主要参考官网Point cloud classification with PointNet
当前环境是python3.6,所以按照博客中方法创建一个使用python3.8的虚拟环境,并进入虚拟环境下完成一些预库的安装。

安装下trimesh库
所安装的tensorflow库是tensorflow-gpu 2.4, cuda版本是11.2, matplotlib3.1.3。若pip安装超时失败,可以手动去网址上下载。Simple Index![]() https://pypi.tuna.tsinghua.edu.cn/simple/比如要下载matplotlib,则可在上述网址基础上跟/matplotlib,即全网址Links for matplotlib
https://pypi.tuna.tsinghua.edu.cn/simple/比如要下载matplotlib,则可在上述网址基础上跟/matplotlib,即全网址Links for matplotlib
运行如下脚本时报错
800
File "/home/sxhlvye/anaconda3/envs/pointnet/lib/python3.8/site-packages/trimesh/graph.py", line 478, in connected_components
raise ImportError('no graph engines available!')
ImportError: no graph engines available!可参考stackflow上的回答error using trimesh library on python - " no graph engine available " - Stack Overflow
完毕后能正常运行,结果如下

第一次modelnet.zip下载完毕后,可以修改下代码,避免每次运行都要重新下载
# This is a sample Python script.
# Press Shift+F10 to execute it or replace it with your code.
# Press Double Shift to search everywhere for classes, files, tool windows, actions, and settings.
import os
import glob
import trimesh
import numpy as np
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from matplotlib import pyplot as plt
tf.random.set_seed(1234)
# DATA_DIR = tf.keras.utils.get_file(
# "modelnet.zip",
# "http://3dvision.princeton.edu/projects/2014/3DShapeNets/ModelNet10.zip",
# extract=True,
# )
# DATA_DIR = os.path.join(os.path.dirname(DATA_DIR), "ModelNet10")
# print(DATA_DIR)
DATA_DIR = "/home/sxhlvye/.keras/datasets/ModelNet10/"
mesh = trimesh.load(os.path.join(DATA_DIR, "chair/train/chair_0001.off"))
mesh.show()这里博主改写下官网示例代码,让其训练的时候可以保存下来模型,供后面的预测阶段来用。代码如下:
# This is a sample Python script.
# Press Shift+F10 to execute it or replace it with your code.
# Press Double Shift to search everywhere for classes, files, tool windows, actions, and settings.
import os
import glob
import trimesh
import numpy as np
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping
tf.random.set_seed(1234)
# DATA_DIR = tf.keras.utils.get_file(
# "modelnet.zip",
# "http://3dvision.princeton.edu/projects/2014/3DShapeNets/ModelNet10.zip",
# extract=True,
# )
# DATA_DIR = os.path.join(os.path.dirname(DATA_DIR), "ModelNet10")
# print(DATA_DIR)
NUM_POINTS = 2048
NUM_CLASSES = 10
BATCH_SIZE = 32
DATA_DIR = "/home/sxhlvye/.keras/datasets/ModelNet10/"
def parse_dataset(num_points=2048):
train_points = []
train_labels = []
test_points = []
test_labels = []
class_map = {}
folders = glob.glob(os.path.join(DATA_DIR, "[!README]*"))
for i, folder in enumerate(folders):
print("processing class: {}".format(os.path.basename(folder)))
# store folder name with ID so we can retrieve later
class_map[i] = folder.split("/")[-1]
# gather all files
train_files = glob.glob(os.path.join(folder, "train/*"))
test_files = glob.glob(os.path.join(folder, "test/*"))
for f in train_files:
train_points.append(trimesh.load(f).sample(num_points))
train_labels.append(i)
for f in test_files:
test_points.append(trimesh.load(f).sample(num_points))
test_labels.append(i)
return (
np.array(train_points),
np.array(test_points),
np.array(train_labels),
np.array(test_labels),
class_map,
)
def augment(points, label):
# jitter points
points += tf.random.uniform(points.shape, -0.005, 0.005, dtype=tf.float64)
# shuffle points
points = tf.random.shuffle(points)
return points, label
def conv_bn(x, filters):
x = layers.Conv1D(filters, kernel_size=1, padding="valid")(x)
x = layers.BatchNormalization(momentum=0.0)(x)
return layers.Activation("relu")(x)
def dense_bn(x, filters):
x = layers.Dense(filters)(x)
x = layers.BatchNormalization(momentum=0.0)(x)
return layers.Activation("relu")(x)
class OrthogonalRegularizer(keras.regularizers.Regularizer):
def __init__(self, num_features, l2reg=0.001):
self.num_features = num_features
self.l2reg = l2reg
self.eye = tf.eye(num_features)
def __call__(self, x):
x = tf.reshape(x, (-1, self.num_features, self.num_features))
xxt = tf.tensordot(x, x, axes=(2, 2))
xxt = tf.reshape(xxt, (-1, self.num_features, self.num_features))
return tf.reduce_sum(self.l2reg * tf.square(xxt - self.eye))
def tnet(inputs, num_features):
# Initalise bias as the indentity matrix
bias = keras.initializers.Constant(np.eye(num_features).flatten())
reg = OrthogonalRegularizer(num_features)
x = conv_bn(inputs, 32)
x = conv_bn(x, 64)
x = conv_bn(x, 512)
x = layers.GlobalMaxPooling1D()(x)
x = dense_bn(x, 256)
x = dense_bn(x, 128)
x = layers.Dense(
num_features * num_features,
kernel_initializer="zeros",
bias_initializer=bias,
activity_regularizer=reg,
)(x)
feat_T = layers.Reshape((num_features, num_features))(x)
# Apply affine transformation to input features
return layers.Dot(axes=(2, 1))([inputs, feat_T])
def testOneImage():
# load one image
mesh = trimesh.load(os.path.join(DATA_DIR, "chair/train/chair_0001.off"))
mesh.show()
points = mesh.sample(2048)
fig = plt.figure(figsize=(5, 5))
ax = fig.add_subplot(111, projection="3d")
ax.scatter(points[:, 0], points[:, 1], points[:, 2])
ax.set_axis_off()
plt.show()
def train(train_dataset, test_dataset):
inputs = keras.Input(shape=(NUM_POINTS, 3))
x = tnet(inputs, 3)
x = conv_bn(x, 32)
x = conv_bn(x, 32)
x = tnet(x, 32)
x = conv_bn(x, 32)
x = conv_bn(x, 64)
x = conv_bn(x, 512)
x = layers.GlobalMaxPooling1D()(x)
x = dense_bn(x, 256)
x = layers.Dropout(0.3)(x)
x = dense_bn(x, 128)
x = layers.Dropout(0.3)(x)
outputs = layers.Dense(NUM_CLASSES, activation="softmax")(x)
model = keras.Model(inputs=inputs, outputs=outputs, name="pointnet")
model.summary()
model.compile(
loss="sparse_categorical_crossentropy",
optimizer=keras.optimizers.Adam(learning_rate=0.001),
metrics=["sparse_categorical_accuracy"],
)
model_checkpoint = ModelCheckpoint("weights.h5", monitor='val_loss', save_best_only=True, save_weights_only=True)
easy_stopping = EarlyStopping(monitor='val_loss', patience=10, verbose=0, mode='min')
history = model.fit(train_dataset, epochs=500, validation_data=test_dataset, callbacks=[model_checkpoint, easy_stopping])
plt.clf()
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.savefig('val_loss.png')
def predict(test_dataset):
data = test_dataset.take(1)
points, labels = list(data)[0]
points = points[:8, ...]
labels = labels[:8, ...]
# run test data through model
inputs = keras.Input(shape=(NUM_POINTS, 3))
x = tnet(inputs, 3)
x = conv_bn(x, 32)
x = conv_bn(x, 32)
x = tnet(x, 32)
x = conv_bn(x, 32)
x = conv_bn(x, 64)
x = conv_bn(x, 512)
x = layers.GlobalMaxPooling1D()(x)
x = dense_bn(x, 256)
x = layers.Dropout(0.3)(x)
x = dense_bn(x, 128)
x = layers.Dropout(0.3)(x)
outputs = layers.Dense(NUM_CLASSES, activation="softmax")(x)
model = keras.Model(inputs=inputs, outputs=outputs, name="pointnet")
model.summary()
model.compile(
loss="sparse_categorical_crossentropy",
optimizer=keras.optimizers.Adam(learning_rate=0.001),
metrics=["sparse_categorical_accuracy"],
)
model.load_weights("weights.h5")
preds = model.predict(points)
preds = tf.math.argmax(preds, -1)
points = points.numpy()
# plot points with predicted class and label
fig = plt.figure(figsize=(15, 10))
for i in range(8):
ax = fig.add_subplot(2, 4, i + 1, projection="3d")
ax.scatter(points[i, :, 0], points[i, :, 1], points[i, :, 2])
ax.set_title(
"pred: {:}, label: {:}".format(
CLASS_MAP[preds[i].numpy()], CLASS_MAP[labels.numpy()[i]]
)
)
ax.set_axis_off()
plt.show()
if __name__=="__main__":
#testOneImage()
#load data
train_points, test_points, train_labels, test_labels, CLASS_MAP = parse_dataset(NUM_POINTS)
print(train_points.shape)
print(test_points.shape)
print(train_labels.shape)
print(test_labels.shape)
train_dataset = tf.data.Dataset.from_tensor_slices((train_points, train_labels))
test_dataset = tf.data.Dataset.from_tensor_slices((test_points, test_labels))
train_dataset = train_dataset.shuffle(len(train_points)).map(augment).batch(BATCH_SIZE)
test_dataset = test_dataset.shuffle(len(test_points)).batch(BATCH_SIZE)
print(train_points.size);
print(test_points.size);
#train data to get a model
#train(train_dataset, test_dataset)
predict(test_dataset)
训练结果如下:
/home/sxhlvye/anaconda3/envs/pointnet/bin/python3 /home/sxhlvye/Trial/keras_pointnet/main.py
2023-05-12 22:18:24.628693: I tensorflow/stream_executor/platform/default/dso_loader.cc:49] Successfully opened dynamic library libcudart.so.11.0
processing class: night_stand
processing class: bed
processing class: desk
processing class: monitor
processing class: sofa
processing class: bathtub
processing class: toilet
processing class: dresser
processing class: table
processing class: chair
2023-05-12 22:22:48.659849: I tensorflow/compiler/jit/xla_cpu_device.cc:41] Not creating XLA devices, tf_xla_enable_xla_devices not set
2023-05-12 22:22:48.660604: I tensorflow/stream_executor/platform/default/dso_loader.cc:49] Successfully opened dynamic library libcuda.so.1
(3991, 2048, 3)
(908, 2048, 3)
(3991,)
(908,)
2023-05-12 22:22:48.697202: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:941] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero
2023-05-12 22:22:48.697567: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1720] Found device 0 with properties:
pciBusID: 0000:01:00.0 name: NVIDIA GeForce GTX 1660 Ti with Max-Q Design computeCapability: 7.5
coreClock: 1.335GHz coreCount: 24 deviceMemorySize: 5.80GiB deviceMemoryBandwidth: 268.26GiB/s
2023-05-12 22:22:48.697591: I tensorflow/stream_executor/platform/default/dso_loader.cc:49] Successfully opened dynamic library libcudart.so.11.0
2023-05-12 22:22:48.699881: I tensorflow/stream_executor/platform/default/dso_loader.cc:49] Successfully opened dynamic library libcublas.so.11
2023-05-12 22:22:48.699919: I tensorflow/stream_executor/platform/default/dso_loader.cc:49] Successfully opened dynamic library libcublasLt.so.11
2023-05-12 22:22:48.700656: I tensorflow/stream_executor/platform/default/dso_loader.cc:49] Successfully opened dynamic library libcufft.so.10
2023-05-12 22:22:48.700867: I tensorflow/stream_executor/platform/default/dso_loader.cc:49] Successfully opened dynamic library libcurand.so.10
2023-05-12 22:22:48.701004: W tensorflow/stream_executor/platform/default/dso_loader.cc:60] Could not load dynamic library 'libcusolver.so.10'; dlerror: libcusolver.so.10: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /home/sxhlvye/Downloads/TensorRT-8.0.0.3/lib:/usr/local/cuda-11.0/lib64
2023-05-12 22:22:48.701459: I tensorflow/stream_executor/platform/default/dso_loader.cc:49] Successfully opened dynamic library libcusparse.so.11
2023-05-12 22:22:48.701586: I tensorflow/stream_executor/platform/default/dso_loader.cc:49] Successfully opened dynamic library libcudnn.so.8
2023-05-12 22:22:48.701596: W tensorflow/core/common_runtime/gpu/gpu_device.cc:1757] Cannot dlopen some GPU libraries. Please make sure the missing libraries mentioned above are installed properly if you would like to use GPU. Follow the guide at https://www.tensorflow.org/install/gpu for how to download and setup the required libraries for your platform.
Skipping registering GPU devices...
2023-05-12 22:22:48.702140: I tensorflow/compiler/jit/xla_gpu_device.cc:99] Not creating XLA devices, tf_xla_enable_xla_devices not set
2023-05-12 22:22:48.702155: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1261] Device interconnect StreamExecutor with strength 1 edge matrix:
2023-05-12 22:22:48.702161: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1267]
24520704
5578752
Model: "pointnet"
__________________________________________________________________________________________________
Layer (type) Output Shape Param # Connected to
==================================================================================================
input_1 (InputLayer) [(None, 2048, 3)] 0
__________________________________________________________________________________________________
conv1d (Conv1D) (None, 2048, 32) 128 input_1[0][0]
__________________________________________________________________________________________________
batch_normalization (BatchNorma (None, 2048, 32) 128 conv1d[0][0]
__________________________________________________________________________________________________
activation (Activation) (None, 2048, 32) 0 batch_normalization[0][0]
__________________________________________________________________________________________________
conv1d_1 (Conv1D) (None, 2048, 64) 2112 activation[0][0]
__________________________________________________________________________________________________
batch_normalization_1 (BatchNor (None, 2048, 64) 256 conv1d_1[0][0]
__________________________________________________________________________________________________
activation_1 (Activation) (None, 2048, 64) 0 batch_normalization_1[0][0]
__________________________________________________________________________________________________
conv1d_2 (Conv1D) (None, 2048, 512) 33280 activation_1[0][0]
__________________________________________________________________________________________________
batch_normalization_2 (BatchNor (None, 2048, 512) 2048 conv1d_2[0][0]
__________________________________________________________________________________________________
activation_2 (Activation) (None, 2048, 512) 0 batch_normalization_2[0][0]
__________________________________________________________________________________________________
global_max_pooling1d (GlobalMax (None, 512) 0 activation_2[0][0]
__________________________________________________________________________________________________
dense (Dense) (None, 256) 131328 global_max_pooling1d[0][0]
__________________________________________________________________________________________________
batch_normalization_3 (BatchNor (None, 256) 1024 dense[0][0]
__________________________________________________________________________________________________
activation_3 (Activation) (None, 256) 0 batch_normalization_3[0][0]
__________________________________________________________________________________________________
dense_1 (Dense) (None, 128) 32896 activation_3[0][0]
__________________________________________________________________________________________________
batch_normalization_4 (BatchNor (None, 128) 512 dense_1[0][0]
__________________________________________________________________________________________________
activation_4 (Activation) (None, 128) 0 batch_normalization_4[0][0]
__________________________________________________________________________________________________
dense_2 (Dense) (None, 9) 1161 activation_4[0][0]
__________________________________________________________________________________________________
reshape (Reshape) (None, 3, 3) 0 dense_2[0][0]
__________________________________________________________________________________________________
dot (Dot) (None, 2048, 3) 0 input_1[0][0]
reshape[0][0]
__________________________________________________________________________________________________
conv1d_3 (Conv1D) (None, 2048, 32) 128 dot[0][0]
__________________________________________________________________________________________________
batch_normalization_5 (BatchNor (None, 2048, 32) 128 conv1d_3[0][0]
__________________________________________________________________________________________________
activation_5 (Activation) (None, 2048, 32) 0 batch_normalization_5[0][0]
__________________________________________________________________________________________________
conv1d_4 (Conv1D) (None, 2048, 32) 1056 activation_5[0][0]
__________________________________________________________________________________________________
batch_normalization_6 (BatchNor (None, 2048, 32) 128 conv1d_4[0][0]
__________________________________________________________________________________________________
activation_6 (Activation) (None, 2048, 32) 0 batch_normalization_6[0][0]
__________________________________________________________________________________________________
conv1d_5 (Conv1D) (None, 2048, 32) 1056 activation_6[0][0]
__________________________________________________________________________________________________
batch_normalization_7 (BatchNor (None, 2048, 32) 128 conv1d_5[0][0]
__________________________________________________________________________________________________
activation_7 (Activation) (None, 2048, 32) 0 batch_normalization_7[0][0]
__________________________________________________________________________________________________
conv1d_6 (Conv1D) (None, 2048, 64) 2112 activation_7[0][0]
__________________________________________________________________________________________________
batch_normalization_8 (BatchNor (None, 2048, 64) 256 conv1d_6[0][0]
__________________________________________________________________________________________________
activation_8 (Activation) (None, 2048, 64) 0 batch_normalization_8[0][0]
__________________________________________________________________________________________________
conv1d_7 (Conv1D) (None, 2048, 512) 33280 activation_8[0][0]
__________________________________________________________________________________________________
batch_normalization_9 (BatchNor (None, 2048, 512) 2048 conv1d_7[0][0]
__________________________________________________________________________________________________
activation_9 (Activation) (None, 2048, 512) 0 batch_normalization_9[0][0]
__________________________________________________________________________________________________
global_max_pooling1d_1 (GlobalM (None, 512) 0 activation_9[0][0]
__________________________________________________________________________________________________
dense_3 (Dense) (None, 256) 131328 global_max_pooling1d_1[0][0]
__________________________________________________________________________________________________
batch_normalization_10 (BatchNo (None, 256) 1024 dense_3[0][0]
__________________________________________________________________________________________________
activation_10 (Activation) (None, 256) 0 batch_normalization_10[0][0]
__________________________________________________________________________________________________
dense_4 (Dense) (None, 128) 32896 activation_10[0][0]
__________________________________________________________________________________________________
batch_normalization_11 (BatchNo (None, 128) 512 dense_4[0][0]
__________________________________________________________________________________________________
activation_11 (Activation) (None, 128) 0 batch_normalization_11[0][0]
__________________________________________________________________________________________________
dense_5 (Dense) (None, 1024) 132096 activation_11[0][0]
__________________________________________________________________________________________________
reshape_1 (Reshape) (None, 32, 32) 0 dense_5[0][0]
__________________________________________________________________________________________________
dot_1 (Dot) (None, 2048, 32) 0 activation_6[0][0]
reshape_1[0][0]
__________________________________________________________________________________________________
conv1d_8 (Conv1D) (None, 2048, 32) 1056 dot_1[0][0]
__________________________________________________________________________________________________
batch_normalization_12 (BatchNo (None, 2048, 32) 128 conv1d_8[0][0]
__________________________________________________________________________________________________
activation_12 (Activation) (None, 2048, 32) 0 batch_normalization_12[0][0]
__________________________________________________________________________________________________
conv1d_9 (Conv1D) (None, 2048, 64) 2112 activation_12[0][0]
__________________________________________________________________________________________________
batch_normalization_13 (BatchNo (None, 2048, 64) 256 conv1d_9[0][0]
__________________________________________________________________________________________________
activation_13 (Activation) (None, 2048, 64) 0 batch_normalization_13[0][0]
__________________________________________________________________________________________________
conv1d_10 (Conv1D) (None, 2048, 512) 33280 activation_13[0][0]
__________________________________________________________________________________________________
batch_normalization_14 (BatchNo (None, 2048, 512) 2048 conv1d_10[0][0]
__________________________________________________________________________________________________
activation_14 (Activation) (None, 2048, 512) 0 batch_normalization_14[0][0]
__________________________________________________________________________________________________
global_max_pooling1d_2 (GlobalM (None, 512) 0 activation_14[0][0]
__________________________________________________________________________________________________
dense_6 (Dense) (None, 256) 131328 global_max_pooling1d_2[0][0]
__________________________________________________________________________________________________
batch_normalization_15 (BatchNo (None, 256) 1024 dense_6[0][0]
__________________________________________________________________________________________________
activation_15 (Activation) (None, 256) 0 batch_normalization_15[0][0]
__________________________________________________________________________________________________
dropout (Dropout) (None, 256) 0 activation_15[0][0]
__________________________________________________________________________________________________
dense_7 (Dense) (None, 128) 32896 dropout[0][0]
__________________________________________________________________________________________________
batch_normalization_16 (BatchNo (None, 128) 512 dense_7[0][0]
__________________________________________________________________________________________________
activation_16 (Activation) (None, 128) 0 batch_normalization_16[0][0]
__________________________________________________________________________________________________
dropout_1 (Dropout) (None, 128) 0 activation_16[0][0]
__________________________________________________________________________________________________
dense_8 (Dense) (None, 10) 1290 dropout_1[0][0]
==================================================================================================
Total params: 748,979
Trainable params: 742,899
Non-trainable params: 6,080
__________________________________________________________________________________________________
2023-05-12 22:22:49.304417: I tensorflow/compiler/mlir/mlir_graph_optimization_pass.cc:116] None of the MLIR optimization passes are enabled (registered 2)
2023-05-12 22:22:49.322971: I tensorflow/core/platform/profile_utils/cpu_utils.cc:112] CPU Frequency: 2599990000 Hz
Epoch 1/500
125/125 [==============================] - 284s 2s/step - loss: 4.0342 - sparse_categorical_accuracy: 0.2180 - val_loss: 20163459072.0000 - val_sparse_categorical_accuracy: 0.2026
Epoch 2/500
125/125 [==============================] - 283s 2s/step - loss: 3.0899 - sparse_categorical_accuracy: 0.3759 - val_loss: 4261103009792.0000 - val_sparse_categorical_accuracy: 0.3744
Epoch 3/500
125/125 [==============================] - 283s 2s/step - loss: 2.7776 - sparse_categorical_accuracy: 0.4847 - val_loss: 38475904.0000 - val_sparse_categorical_accuracy: 0.4570
Epoch 4/500
125/125 [==============================] - 283s 2s/step - loss: 2.4536 - sparse_categorical_accuracy: 0.5812 - val_loss: 308573824.0000 - val_sparse_categorical_accuracy: 0.5055
Epoch 5/500
125/125 [==============================] - 283s 2s/step - loss: 2.2765 - sparse_categorical_accuracy: 0.6288 - val_loss: 1299249408.0000 - val_sparse_categorical_accuracy: 0.5969
Epoch 6/500
125/125 [==============================] - 284s 2s/step - loss: 2.2702 - sparse_categorical_accuracy: 0.6390 - val_loss: 2634373488705536.0000 - val_sparse_categorical_accuracy: 0.7467
Epoch 7/500
125/125 [==============================] - 284s 2s/step - loss: 2.0334 - sparse_categorical_accuracy: 0.7036 - val_loss: 2378090035910948159488.0000 - val_sparse_categorical_accuracy: 0.6189
Epoch 8/500
125/125 [==============================] - 285s 2s/step - loss: 2.0664 - sparse_categorical_accuracy: 0.6943 - val_loss: 854828.3750 - val_sparse_categorical_accuracy: 0.6916
Epoch 9/500
125/125 [==============================] - 285s 2s/step - loss: 1.9842 - sparse_categorical_accuracy: 0.7237 - val_loss: 3.5205 - val_sparse_categorical_accuracy: 0.2214
Epoch 10/500
125/125 [==============================] - 284s 2s/step - loss: 1.8826 - sparse_categorical_accuracy: 0.7645 - val_loss: 4862585775792848896.0000 - val_sparse_categorical_accuracy: 0.7026
Epoch 11/500
125/125 [==============================] - 283s 2s/step - loss: 1.7948 - sparse_categorical_accuracy: 0.7728 - val_loss: 3543.4460 - val_sparse_categorical_accuracy: 0.6795
Epoch 12/500
125/125 [==============================] - 283s 2s/step - loss: 1.7943 - sparse_categorical_accuracy: 0.7849 - val_loss: 3.1022 - val_sparse_categorical_accuracy: 0.4747
Epoch 13/500
125/125 [==============================] - 283s 2s/step - loss: 1.7282 - sparse_categorical_accuracy: 0.7975 - val_loss: 11759.7402 - val_sparse_categorical_accuracy: 0.7434
Epoch 14/500
125/125 [==============================] - 284s 2s/step - loss: 1.7557 - sparse_categorical_accuracy: 0.7909 - val_loss: 17441380.0000 - val_sparse_categorical_accuracy: 0.7588
Epoch 15/500
125/125 [==============================] - 284s 2s/step - loss: 1.7371 - sparse_categorical_accuracy: 0.7875 - val_loss: 644425952788480.0000 - val_sparse_categorical_accuracy: 0.7885
Epoch 16/500
125/125 [==============================] - 284s 2s/step - loss: 1.6748 - sparse_categorical_accuracy: 0.8142 - val_loss: 14097967778157821952.0000 - val_sparse_categorical_accuracy: 0.7126
Epoch 17/500
125/125 [==============================] - 283s 2s/step - loss: 1.6334 - sparse_categorical_accuracy: 0.8174 - val_loss: 1968121.7500 - val_sparse_categorical_accuracy: 0.8205
Epoch 18/500
125/125 [==============================] - 283s 2s/step - loss: 1.6253 - sparse_categorical_accuracy: 0.8261 - val_loss: 2.2097 - val_sparse_categorical_accuracy: 0.6542
Epoch 19/500
125/125 [==============================] - 284s 2s/step - loss: 1.5634 - sparse_categorical_accuracy: 0.8458 - val_loss: 167856331292672.0000 - val_sparse_categorical_accuracy: 0.8117
Epoch 20/500
125/125 [==============================] - 283s 2s/step - loss: 1.5733 - sparse_categorical_accuracy: 0.8477 - val_loss: 5170860032.0000 - val_sparse_categorical_accuracy: 0.8084
Epoch 21/500
125/125 [==============================] - 285s 2s/step - loss: 1.5365 - sparse_categorical_accuracy: 0.8484 - val_loss: 3753467707392.0000 - val_sparse_categorical_accuracy: 0.8458
Epoch 22/500
125/125 [==============================] - 285s 2s/step - loss: 1.5660 - sparse_categorical_accuracy: 0.8428 - val_loss: 3.1326 - val_sparse_categorical_accuracy: 0.6894
Epoch 23/500
125/125 [==============================] - 285s 2s/step - loss: 1.5872 - sparse_categorical_accuracy: 0.8314 - val_loss: 121918711660544.0000 - val_sparse_categorical_accuracy: 0.8513
Epoch 24/500
125/125 [==============================] - 283s 2s/step - loss: 1.4560 - sparse_categorical_accuracy: 0.8758 - val_loss: 1849604736.0000 - val_sparse_categorical_accuracy: 0.7753
Epoch 25/500
125/125 [==============================] - 283s 2s/step - loss: 1.4628 - sparse_categorical_accuracy: 0.8741 - val_loss: 95.5576 - val_sparse_categorical_accuracy: 0.7874
Epoch 26/500
125/125 [==============================] - 283s 2s/step - loss: 1.4370 - sparse_categorical_accuracy: 0.8845 - val_loss: 404704232210432.0000 - val_sparse_categorical_accuracy: 0.7599
Epoch 27/500
125/125 [==============================] - 283s 2s/step - loss: 1.4637 - sparse_categorical_accuracy: 0.8685 - val_loss: 340569760.0000 - val_sparse_categorical_accuracy: 0.8579
Epoch 28/500
125/125 [==============================] - 284s 2s/step - loss: 1.4841 - sparse_categorical_accuracy: 0.8661 - val_loss: 567234592768.0000 - val_sparse_categorical_accuracy: 0.8601
Process finished with exit code 0迭代损失率曲线如下:

预测结果如下:

后续还需要再优化下参数。这个例子只是拿pointNet网络做点云的分类,并没有展示语义分割的功能。
PointNet的相关理论知识可参考论文https://openaccess.thecvf.com/content_cvpr_2017/papers/Qi_PointNet_Deep_Learning_CVPR_2017_paper.pdf
也可以参考一些博客的详解
Pointnet以及Pointnet++论文笔记_慢下去、静下来的博客-CSDN博客
搞懂PointNet++,这篇文章就够了! - 知乎
2. 3D convolutional networks(Openvino实现)
可参考open_model_zoo/demos/3d_segmentation_demo/python at master · openvinotoolkit/open_model_zoo · GitHub
Openvino的详细配置可参考博主之前博客,这边不细说了,只会贴上一些记录过程。
conda create -n test3DCNN python=3.7
source activate test3DCNN
python -m pip install openvino-dev[onnx,pytorch,mxnet]
下载过程中,opencv_python库会因为超时而报错,可从如下链接处下载
Links for opencv-python
完毕后,下载demo示例到本地磁盘上
使用如下命令语句下载模型
omz_downloader --list models.lst会看到模型已经下载到如下位置了

再使用 如下命令语句进行转换
omz_converter --list models.lst转换后的模型如下
这里用pycharm社区版来打开工程
模型有了,那还差测试数据,可从如下官网上下载
Results![]() http://medicaldecathlon.com/dataaws/关于数据集的介绍,也可以从这里获取
http://medicaldecathlon.com/dataaws/关于数据集的介绍,也可以从这里获取
3-D Brain Tumor Segmentation Using Deep Learning- MATLAB & Simulink Example
下载完毕后,拿训练集中的BRATS_001.nii单个文件来做测试,同时也一张张保存了对应CT图像的每层的标注图。工程目录结构如下:

3d_segmentation_demo.py文件中代码如下:
#!/usr/bin/env python3
"""
Copyright (c) 2019-2023 Intel Corporation
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
"""
import os
import sys
import logging as log
from time import perf_counter
import numpy as np
import nibabel as nib
from PIL import Image, ImageSequence
from argparse import ArgumentParser, SUPPRESS
from fnmatch import fnmatch
from scipy.ndimage import interpolation
from openvino.runtime import Core, get_version, PartialShape
import os
import cv2
print(os.getcwd())
log.basicConfig(format='[ %(levelname)s ] %(message)s', level=log.DEBUG, stream=sys.stdout)
CLASSES_COLOR_MAP = [
(150, 150, 150),
(58, 55, 169),
(211, 51, 17),
(157, 80, 44),
(23, 95, 189),
(210, 133, 34),
(76, 226, 202),
(101, 138, 127),
(223, 91, 182),
(80, 128, 113),
(235, 155, 55),
(44, 151, 243),
(159, 80, 170),
(239, 208, 44),
(128, 50, 51),
(82, 141, 193),
(9, 107, 10),
(223, 90, 142),
(50, 248, 83),
(178, 101, 130),
(71, 30, 204)
]
# suffixes for original interpretation in dataset
SUFFIX_T1 = "_t1.nii.gz"
SUFFIX_T2 = "_t2.nii.gz"
SUFFIX_FLAIR = "_flair.nii.gz"
SUFFIX_T1CE = "_t1ce.nii.gz"
SUFFIX_SEG = "_seg.nii.gz"
# file suffixes to form a data tensor
DATA_SUFFIXES = [SUFFIX_T1, SUFFIX_T2, SUFFIX_FLAIR, SUFFIX_T1CE]
NIFTI_FOLDER = 0
NIFTI_FILE = 1
TIFF_FILE = 2
def mri_sequence(arg):
sequence = tuple(int(k) for k in arg.split(','))
if len(sequence) != 4:
raise AttributeError("The MRI-sequence should contain exactly 4 values, but contains {}.".format(len(sequence)))
if len(set(sequence)) != 4:
raise AttributeError("The MRI-sequence has repeating scan types - {}. "
"The MRI-sequence must contain native T1, native T2, T2-FLAIR, "
"post-Gadolinium contrast T1 scans in the specific for the net order".
format(sequence))
return sequence
def parse_arguments():
parser = ArgumentParser(add_help=False)
args = parser.add_argument_group('Options')
args.add_argument('-h', '--help', action='help', default=SUPPRESS, help='Show this help message and exit.')
args.add_argument('-i', '--path_to_input_data', type=str, required=False, default="/home/sxhlvye/open_model_zoo-master/demos/3d_segmentation_demo/python/BRATS_001.nii",
help="Required. Path to an input folder with NIfTI data/NIFTI file/TIFF file")
args.add_argument('-m', '--path_to_model', type=str, required=False,default="./public/brain-tumor-segmentation-0001/FP32/brain-tumor-segmentation-0001.xml",
help="Required. Path to an .xml file with a trained model")
args.add_argument('-o', '--path_to_output', type=str, required=False, default="./result",
help="Required. Path to a folder where output files will be saved")
args.add_argument('-d', '--target_device', type=str, required=False, default="GPU",
help="Optional. Specify a target device to infer on: CPU, GPU. "
"Use \"-d HETERO:<comma separated devices list>\" format to specify HETERO plugin.")
args.add_argument("-nii", "--output_nifti", help="Show output inference results as raw values", default=False,
action="store_true")
args.add_argument('-nthreads', '--number_threads', type=int, required=False, default=None,
help="Optional. Number of threads to use for inference on CPU (including HETERO cases).")
args.add_argument('-s', '--shape', nargs='*', type=int, required=False, default=None,
help="Optional. Specify shape for a network")
args.add_argument('-ms', '--mri_sequence', type=mri_sequence, metavar='N1,N2,N3,N4', default=(2, 0, 3, 1),
help='Optional. Transfer MRI-sequence from dataset order to the network order.')
args.add_argument("--full_intensities_range", required=False, default=False, action="store_true",
help="Take intensities of the input image in a full range.")
return parser.parse_args()
def get_input_type(path):
if os.path.isdir(path):
return NIFTI_FOLDER
elif fnmatch(path, '*.nii.gz') or fnmatch(path, '*.nii'):
return NIFTI_FILE
elif fnmatch(path, '*.tif') or fnmatch(path, '*.tiff'):
return TIFF_FILE
raise RuntimeError("Input must be a folder with 4 NIFTI files, single NIFTI file (*.nii or *.nii.gz) or "
"TIFF file (*.tif or *.tiff)")
def find_series_name(path):
for file in os.listdir(path):
if fnmatch(file, '*.nii.gz'):
for suffix in DATA_SUFFIXES:
if suffix in file:
return file.replace(suffix, '')
def bbox3(img):
rows = np.any(img, axis=1)
rows = np.any(rows, axis=1)
rows = np.where(rows)
cols = np.any(img, axis=0)
cols = np.any(cols, axis=1)
cols = np.where(cols)
slices = np.any(img, axis=0)
slices = np.any(slices, axis=0)
slices = np.where(slices)
if (rows[0].shape[0] > 0):
rmin, rmax = rows[0][[0, -1]]
cmin, cmax = cols[0][[0, -1]]
smin, smax = slices[0][[0, -1]]
return np.array([[rmin, cmin, smin], [rmax, cmax, smax]])
return np.array([[-1, -1, -1], [0, 0, 0]])
def read_nii_header(data_path, name):
filename = os.path.join(data_path, name)
if not os.path.exists(filename):
raise ValueError("File {} is not exist. Please, validate path to input".format(filename))
return nib.load(filename)
def normalize(image, mask, full_intensities_range):
ret = image.copy()
image_masked = np.ma.masked_array(ret, ~(mask))
ret = ret - np.mean(image_masked)
ret = ret / np.var(image_masked) ** 0.5
if not full_intensities_range:
ret[ret > 5.] = 5.
ret[ret < -5.] = -5.
ret += 5.
ret /= 10
ret[~mask] = 0.
return ret
def resample_np(data, output_shape, order):
assert len(data.shape) == len(output_shape)
factor = [float(o) / i for i, o in zip(data.shape, output_shape)]
return interpolation.zoom(data, zoom=factor, order=order)
def read_image(test_data_path, data_name, sizes=(128, 128, 128), is_series=True,
mri_sequence_order=(0, 1, 2, 3), full_intensities_range=False):
images_list = []
original_shape = ()
bboxes = np.zeros(shape=(len(DATA_SUFFIXES),) + (2, 3))
if is_series:
data_seq = [DATA_SUFFIXES[i] for i in mri_sequence_order]
for j, s in enumerate(data_seq):
image_handle = read_nii_header(test_data_path, data_name + s)
affine = image_handle.affine
image = image_handle.get_fdata(dtype=np.float32)
mask = image > 0.
bboxes[j] = bbox3(mask)
image = normalize(image, mask, full_intensities_range)
images_list.append(image.reshape((1, 1,) + image.shape))
original_shape = image.shape
else:
data_handle = read_nii_header(test_data_path, data_name)
affine = data_handle.affine
data = data_handle.get_fdata(dtype=np.float32)
assert len(data.shape) == 4, 'Wrong data dimensions - {}, must be 4'.format(len(data.shape))
assert data.shape[3] == 4, 'Wrong data shape - {}, must be (:,:,:,4)'.format(data.shape)
# Reading order is specified for data from http://medicaldecathlon.com/
for j in mri_sequence_order:
image = data[:, :, :, j]
mask = image > 0
bboxes[j] = bbox3(mask)
image = normalize(image, mask, full_intensities_range)
images_list.append(image.reshape((1, 1,) + image.shape))
original_shape = data.shape[:3]
bbox_min = np.min(bboxes[:, 0, :], axis=0).ravel().astype(int)
bbox_max = np.max(bboxes[:, 1, :], axis=0).ravel().astype(int)
bbox = np.zeros(shape=(2, 3), dtype=float)
bbox[0] = bbox_min
bbox[1] = bbox_max
data = np.concatenate(images_list, axis=1)
if data.shape[2:] == sizes:
data_crop = data
else:
data_crop = resample_np(
data[:, :, bbox_min[0]:bbox_max[0], bbox_min[1]:bbox_max[1], bbox_min[2]:bbox_max[2]],
(1, len(DATA_SUFFIXES),) + sizes,
1)
bbox_ret = [
bbox_min[0], bbox_max[0],
bbox_min[1], bbox_max[1],
bbox_min[2], bbox_max[2]
]
return data, data_crop, affine, original_shape, bbox_ret
def showLabel():
data_handle = read_nii_header("", "label.nii")
affine = data_handle.affine
data = data_handle.get_fdata(dtype=np.float32)
print(data.shape)
for i in range(0, data.shape[2]):
label = np.array(data[:, :,i])
label_color = np.zeros(shape=(data.shape[0],data.shape[1], 3), dtype=np.uint8)
for idx, c in enumerate(CLASSES_COLOR_MAP):
label_color[label[:, :] == idx, :] = np.array(c, dtype=np.uint8)
# cv2.imshow("labelcolor",label_color)
# cv2.waitKey(0)
cv2.imwrite("label/labelcolor" + str(i) + ".bmp", label_color)
def main():
#for check the label
showLabel()
args = parse_arguments()
log.info('OpenVINO Runtime')
log.info('\tbuild: {}'.format(get_version()))
core = Core()
if 'CPU' in args.target_device:
if args.number_threads is not None:
core.set_property("CPU", {'CPU_THREADS_NUM': str(args.number_threads)})
elif 'GPU' not in args.target_device:
raise AttributeError("Device {} do not support of 3D convolution. "
"Please use CPU, GPU or HETERO:*CPU*, HETERO:*GPU*")
log.info('Reading model {}'.format(args.path_to_model))
model = core.read_model(args.path_to_model)
if len(model.inputs) != 1:
raise RuntimeError("only 1 input layer model is supported")
input_tensor_name = model.inputs[0].get_any_name()
if args.shape:
log.debug("Reshape model from {} to {}".format(model.inputs[0].shape, args.shape))
model.reshape({input_tensor_name: PartialShape(args.shape)})
if len(model.inputs[0].shape) != 5:
raise RuntimeError("Incorrect shape {} for 3d convolution network".format(args.shape))
n, c, d, h, w = model.inputs[0].shape
compiled_model = core.compile_model(model, args.target_device)
output_tensor = compiled_model.outputs[0]
infer_request = compiled_model.create_infer_request()
log.info('The model {} is loaded to {}'.format(args.path_to_model, args.target_device))
start_time = perf_counter()
if not os.path.exists(args.path_to_input_data):
raise AttributeError("Path to input data: '{}' does not exist".format(args.path_to_input_data))
input_type = get_input_type(args.path_to_input_data)
is_nifti_data = (input_type == NIFTI_FILE or input_type == NIFTI_FOLDER)
if input_type == NIFTI_FOLDER:
series_name = find_series_name(args.path_to_input_data)
original_data, data_crop, affine, original_size, bbox = \
read_image(args.path_to_input_data, data_name=series_name, sizes=(d, h, w),
mri_sequence_order=args.mri_sequence, full_intensities_range=args.full_intensities_range)
elif input_type == NIFTI_FILE:
original_data, data_crop, affine, original_size, bbox = \
read_image(args.path_to_input_data, data_name=args.path_to_input_data, sizes=(d, h, w), is_series=False,
mri_sequence_order=args.mri_sequence, full_intensities_range=args.full_intensities_range)
else:
data_crop = np.zeros(shape=(n, c, d, h, w), dtype=np.float)
im_seq = ImageSequence.Iterator(Image.open(args.path_to_input_data))
for i, page in enumerate(im_seq):
im = np.array(page).reshape(h, w, c)
for channel in range(c):
data_crop[:, channel, i, :, :] = im[:, :, channel]
original_data = data_crop
original_size = original_data.shape[-3:]
input_data = {input_tensor_name: data_crop}
result = infer_request.infer(input_data)[output_tensor]
batch, channels, out_d, out_h, out_w = result.shape
list_img = []
list_seg_result = []
for batch, data in enumerate(result):
seg_result = np.zeros(shape=original_size, dtype=np.uint8)
if data.shape[1:] != original_size:
x = bbox[1] - bbox[0]
y = bbox[3] - bbox[2]
z = bbox[5] - bbox[4]
out_result = np.zeros(shape=((channels,) + original_size), dtype=float)
out_result[:, bbox[0]:bbox[1], bbox[2]:bbox[3], bbox[4]:bbox[5]] = \
resample_np(data, (channels, x, y, z), 1)
else:
out_result = data
if channels == 1:
reshaped_data = out_result.reshape(original_size[0], original_size[1], original_size[2])
mask = reshaped_data[:, :, :] > 0.5
reshaped_data[mask] = 1
seg_result = reshaped_data.astype(int)
elif channels == 4:
seg_result = np.argmax(out_result, axis=0).astype(int)
elif channels == 3:
res = np.zeros(shape=out_result.shape, dtype=bool)
res = out_result > 0.5
wt = res[0]
tc = res[1]
et = res[2]
seg_result[wt] = 2
seg_result[tc] = 1
seg_result[et] = 3
im = np.stack([original_data[batch, 0, :, :, :],
original_data[batch, 0, :, :, :],
original_data[batch, 0, :, :, :]],
axis=3)
im = 255 * (im - im.min())/(im.max() - im.min())
color_seg_frame = np.zeros(im.shape, dtype=np.uint8)
for idx, c in enumerate(CLASSES_COLOR_MAP):
color_seg_frame[seg_result[:, :, :] == idx, :] = np.array(c, dtype=np.uint8)
mask = seg_result[:, :, :] > 0
im[mask] = color_seg_frame[mask]
for k in range(im.shape[2]):
if is_nifti_data:
list_img.append(Image.fromarray(im[:, :, k, :].astype('uint8'), 'RGB'))
if k == 99:
shap0 = im.shape[0]
shape1 = im.shape[1]
shape2 = im.shape[2]
shap3 = im.shape[3]
#
# g = im[:, :, k, :].astype('uint8')
# cv2.imshow("img",g)
# cv2.waitKey(0)
# Image.fromarray(im[:, :, k, :].astype('uint8'), 'RGB').show()
else:
list_img.append(Image.fromarray(im[k, :, :, :].astype('uint8'), 'RGB'))
if args.output_nifti and is_nifti_data:
list_seg_result.append(seg_result)
total_latency = (perf_counter() - start_time) * 1e3
log.info("Metrics report:")
log.info("\tLatency: {:.1f} ms".format(total_latency))
tiff_output_name = os.path.join(args.path_to_output, 'output.tiff')
image = Image.new('RGB', (original_data.shape[3], original_data.shape[2]))
image.save(tiff_output_name, append_images=list_img, save_all=True)
index = 0
for OneImage in list_img:
OneImage.save("./result/" + str(index) + ".bmp")
index = index + 1
log.debug("Result tiff file was saved to {}".format(tiff_output_name))
if args.output_nifti and is_nifti_data:
for seg_res in list_seg_result:
nii_filename = os.path.join(args.path_to_output, 'output_{}.nii.gz'.format(list_seg_result.index(seg_res)))
nib.save(nib.Nifti1Image(seg_res, affine=affine), nii_filename)
log.debug("Result nifti file was saved to {}".format(nii_filename))
if __name__ == "__main__":
sys.exit(main() or 0)
CT每层的预测结果都有保存到磁盘上,可和标注图做对比。
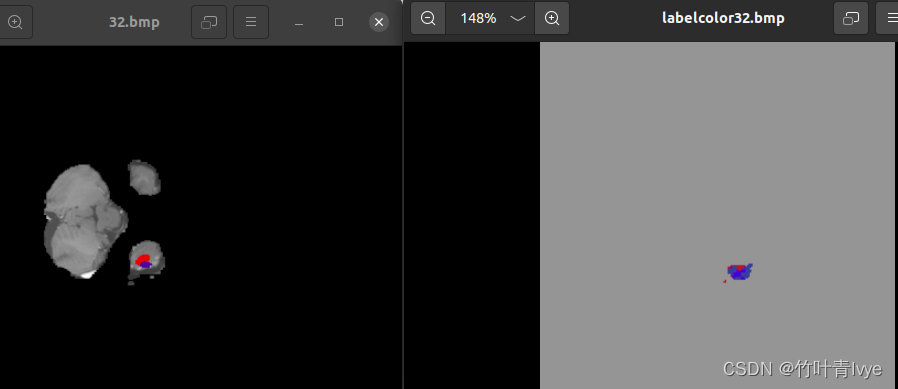
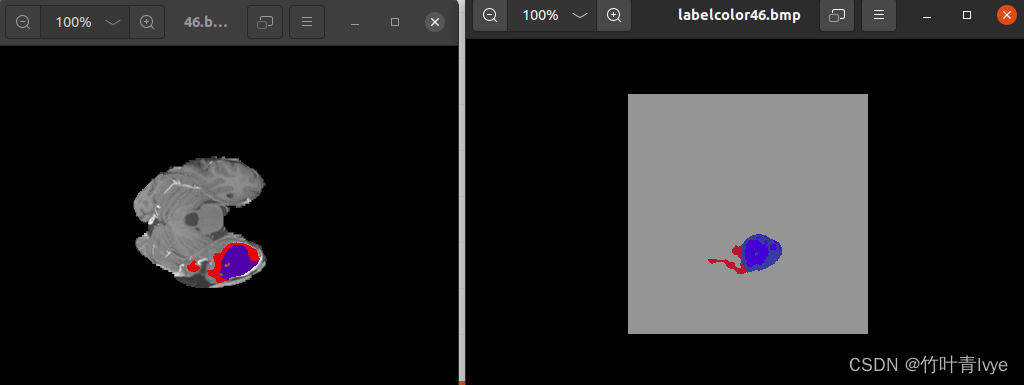
这个示例不涉及模型的训练,只是拿官方的模型来跑跑效果而已,到此就结束了,后续博主会尝试训练自己的点云数据。
3DCNN的一些理论知识可参考论文https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.169.4046&rep=rep1&type=pdf
其它博客帖子可参考如下:
3D-CNN_3dcnn_qinxin_ni的博客-CSDN博客
3DCNN概述及3DCNN与2DCNN的区别_wwz_20的博客-CSDN博客
深度学习笔记----三维卷积及其应用(3DCNN,PointNet,3D U-Net)_YOULANSHENGMENG的博客-CSDN博客
补充(三维点云扫描技术整理):
博主自2017年开始接触到3D点云处理,近几年应用范围越来越广,也越来越火热了。主要是在工业检测、自动驾驶、整形医疗等领域应用越来越广泛了。当年一并接触了深度学习技术和点云处理技术,身边从事该方面工作的童鞋还不多,如今已是火遍整个AI领域了。如下是对三维点云扫描技术的一些整理和一些参考资料的整理。
3D成像方法 汇总(原理解析)--- 双目视觉、激光三角、结构光、ToF、光场、全息 - 知乎
论文《基于数字散斑的双目视觉三维重建方法研究》
【自动驾驶】【快速入门】3D点云与PointNet - 知乎
3D成像技术介绍 - 知乎
自动驾驶(八)---------基于视觉的SLAM_一实相印的博客-CSDN博客
自动驾驶基础知识(一)-什么是自动驾驶 - 知乎
一.结构光
它是利用投影设备将事先设计好的特定模式的结构光图案投影到物体表面,然后使用相机拍摄变形图案,通过分析投影图案在物体表面产生的畸变,再利用投射的特定的编码方案,利用相机针孔成像模式的三角测量原理,得到待测物体上的点的立体坐标。可参考如下博客内容
1. 点结构光
使用半导体激光器将光点投射到被测梧桐表面,通过摄像机获取图像,根据相机成像模型分析光点在相机敏感面和物体表面的位置获得物体的深度信息。需要借助三维位移平台逐点采集和逐次拍照。可参考论文《结构光三维重建技术研究与应用》
2. 线结构光
目前市面上一种很常见的采集技术了,若要形成面,则需要借助于运动平台。主要由线结构光发射器和相机组成,根据激光三角测量原理,由线结构光发射器发射处激光,投射在物体表面上并发生漫反射,反射光进入相机,生成二维图像,通过相机成像之间的关系及预先计算的结构光标定参数,将图像坐标转换为三维点云坐标,同时配合运动平台的移动,可得到被测物体的三维点云数据。详细的一些公式原理推导可以参看论文《基于结构光的软包电池三维重构及表面缺陷检测技术研究》。
3.多线结构光
该方法可以同时处理一幅结构光图像钟的多个特征条纹。这里面提到最多的就是光栅结构光法,可以利用投影装置将光栅条投影到被测物体的表面。
4.编码结构光
编码结构光是为了便于空间中的物点到图像点的映射,将结构光图案按照特定的模式进行设计而来的。编码方法由时间编码方法,空间编码方法,直接编码方法和混合编码方法。通过计算机根据解码原理求出被测对物体表面变形编码图案的解码值,然后利用结构光的基本原理,求出目标上相应的三维坐标。详细的一些公式原理推导可以参看论文《基于编码结构光的三维重建及图像处理研究》
三维重建之结构光编码方案研究_线结构光_少杰很帅的博客-CSDN博客
《基于数字光栅投影结构光的三维重建技术研究》
二.TOF
可参考如下一些资料
TOF技术全解读 - 知乎