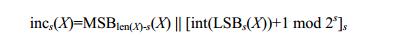DiffDock源码解析
数据预处理
数据输入方式
df = pd.read_csv(args.protein_ligand_csv), 使用的是csv的方式输入,
格式:

不管受体还是配体, 输入可以是序列或者3维结构的文件
如果蛋白输入的是序列,需要计算蛋白的三维结构(ESM模型):
def generate_ESM_structure(model, filename, sequence):
model.set_chunk_size(256)
chunk_size = 256
output = None
while output is None:
with torch.no_grad():
output = model.infer_pdb(sequence)
with open(filename, "w") as f:
f.write(output)
print("saved", filename)
。。。。。。
return output is not None
计算蛋白Embedding
蛋白序列embedding使用了google ESM框架,
def compute_ESM_embeddings(model, alphabet, labels, sequences):
# settings used
toks_per_batch = 4096
repr_layers = [33]
include = "per_tok"
truncation_seq_length = 1022
dataset = FastaBatchedDataset(labels, sequences)
batches = dataset.get_batch_indices(toks_per_batch, extra_toks_per_seq=1)
data_loader = torch.utils.data.DataLoader(
dataset, collate_fn=alphabet.get_batch_converter(truncation_seq_length), batch_sampler=batches
)
assert all(-(model.num_layers + 1) <= i <= model.num_layers for i in repr_layers)
repr_layers = [(i + model.num_layers + 1) % (model.num_layers + 1) for i in repr_layers]
embeddings = {}
with torch.no_grad():
for batch_idx, (labels, strs, toks) in enumerate(data_loader):
print(f"Processing {batch_idx + 1} of {len(batches)} batches ({toks.size(0)} sequences)")
if torch.cuda.is_available():
toks = toks.to(device="cuda", non_blocking=True)
out = model(toks, repr_layers=repr_layers, return_contacts=False)
representations = {layer: t.to(device="cpu") for layer, t in out["representations"].items()}
for i, label in enumerate(labels):
truncate_len = min(truncation_seq_length, len(strs[i]))
embeddings[label] = representations[33][i, 1: truncate_len + 1].clone()
return embeddings

配体预处理
mol = read_molecule(ligand_description, remove_hs=False, sanitize=True)
mol.RemoveAllConformers() # 移除所有的构象信息
mol = AddHs(mol) ## 加氢
generate_conformer(mol) ## 随机3D位置信息
配体特征提取
异构图分子整体信息
complex_graph['ligand'].x = atom_feats
complex_graph['ligand'].pos = lig_coords
complex_graph['ligand', 'lig_bond', 'ligand'].edge_index = edge_index
complex_graph['ligand', 'lig_bond', 'ligand'].edge_attr = edge_attr
- 配体信息特征
allowable_features = {
'possible_atomic_num_list': list(range(1, 119)) + ['misc'],
'possible_chirality_list': [
'CHI_UNSPECIFIED',
'CHI_TETRAHEDRAL_CW',
'CHI_TETRAHEDRAL_CCW',
'CHI_OTHER'
],
'possible_degree_list': [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 'misc'],
'possible_numring_list': [0, 1, 2, 3, 4, 5, 6, 'misc'],
'possible_implicit_valence_list': [0, 1, 2, 3, 4, 5, 6, 'misc'],
'possible_formal_charge_list': [-5, -4, -3, -2, -1, 0, 1, 2, 3, 4, 5, 'misc'],
'possible_numH_list': [0, 1, 2, 3, 4, 5, 6, 7, 8, 'misc'],
'possible_number_radical_e_list': [0, 1, 2, 3, 4, 'misc'],
'possible_hybridization_list': [
'SP', 'SP2', 'SP3', 'SP3D', 'SP3D2', 'misc'
],
'possible_is_aromatic_list': [False, True],
'possible_is_in_ring3_list': [False, True],
'possible_is_in_ring4_list': [False, True],
'possible_is_in_ring5_list': [False, True],
'possible_is_in_ring6_list': [False, True],
'possible_is_in_ring7_list': [False, True],
'possible_is_in_ring8_list': [False, True],
'possible_atom_type_2': ['C*', 'CA', 'CB', 'CD', 'CE', 'CG', 'CH', 'CZ', 'N*', 'ND', 'NE', 'NH', 'NZ', 'O*', 'OD',
'OE', 'OG', 'OH', 'OX', 'S*', 'SD', 'SG', 'misc'],
'possible_atom_type_3': ['C', 'CA', 'CB', 'CD', 'CD1', 'CD2', 'CE', 'CE1', 'CE2', 'CE3', 'CG', 'CG1', 'CG2', 'CH2',
'CZ', 'CZ2', 'CZ3', 'N', 'ND1', 'ND2', 'NE', 'NE1', 'NE2', 'NH1', 'NH2', 'NZ', 'O', 'OD1',
'OD2', 'OE1', 'OE2', 'OG', 'OG1', 'OH', 'OXT', 'SD', 'SG', 'misc'],
}
- 配体位置特征
lig_coords = torch.from_numpy(mol.GetConformer().GetPositions()).float() # 配体的分子位置信息
- 边信息, 键, 无向,键类型作为边属性特征
for bond in mol.GetBonds():
start, end = bond.GetBeginAtomIdx(), bond.GetEndAtomIdx()
row += [start, end]
col += [end, start]
edge_type += 2 * [bonds[bond.GetBondType()]] if bond.GetBondType() != BT.UNSPECIFIED else [0, 0]
受体蛋白特征提取
受体信息提取主要是空间信息,原子空间坐标(coords), 残基CA原子(c_alpha_coords), 残基N原子(n_coords), 残基CB原子(c_coords), 残基嵌入信息(lm_embeddings)
complex_graph['receptor'].x = torch.cat([node_feat, torch.tensor(lm_embeddings)], axis=1) if lm_embeddings is not None else node_feat
complex_graph['receptor'].pos = torch.from_numpy(c_alpha_coords).float()
complex_graph['receptor'].mu_r_norm = mu_r_norm
complex_graph['receptor'].side_chain_vecs = side_chain_vecs.float()
complex_graph['receptor', 'rec_contact', 'receptor'].edge_index = torch.from_numpy(np.asarray([src_list, dst_list])) # 出节点 -> 入节点 cutoff 15.0
- 空间信息特征
n_rel_pos = n_coords - c_alpha_coords # N -> CA
c_rel_pos = c_coords - c_alpha_coords # CB -> CA
mu_r_norm = torch.from_numpy(np.array(mean_norm_list).astype(np.float32))
side_chain_vecs = torch.from_numpy(
np.concatenate([np.expand_dims(n_rel_pos, axis=1), np.expand_dims(c_rel_pos, axis=1)], axis=1))
- 受体信息特征
只使用了残基标签one-hot 与残基序列embedding的拼接
def rec_residue_featurizer(rec):
feature_list = []
for residue in rec.get_residues():
feature_list.append([safe_index(allowable_features['possible_amino_acids'], residue.get_resname())])
return torch.tensor(feature_list, dtype=torch.float32) # (N_res, 1)
'possible_amino_acids': ['ALA', 'ARG', 'ASN', 'ASP', 'CYS', 'GLN', 'GLU', 'GLY', 'HIS', 'ILE', 'LEU', 'LYS', 'MET',
'PHE', 'PRO', 'SER', 'THR', 'TRP', 'TYR', 'VAL', 'HIP', 'HIE', 'TPO', 'HID', 'LEV', 'MEU',
'PTR', 'GLV', 'CYT', 'SEP', 'HIZ', 'CYM', 'GLM', 'ASQ', 'TYS', 'CYX', 'GLZ', 'misc'],
模型
模型有两种,一种是全原子特征, 另一种是CA原子特征
##
def get_model(args, device, t_to_sigma, no_parallel=False, confidence_mode=False):
if 'all_atoms' in args and args.all_atoms:
model_class = AAScoreModel
else:
model_class = CGScoreModel
节点表征
class AtomEncoder(torch.nn.Module):
# feature_dims元组的第一个元素是包含每个分类特征长度的列表,第二个元素是标量特征的数量
def __init__(self, emb_dim, feature_dims, sigma_embed_dim, lm_embedding_type= None):
# first element of feature_dims tuple is a list with the lenght of each categorical feature and the second is the number of scalar features
super(AtomEncoder, self).__init__()
self.atom_embedding_list = torch.nn.ModuleList()
self.num_categorical_features = len(feature_dims[0])
self.num_scalar_features = feature_dims[1] + sigma_embed_dim
self.lm_embedding_type = lm_embedding_type
for i, dim in enumerate(feature_dims[0]):
emb = torch.nn.Embedding(dim, emb_dim)
torch.nn.init.xavier_uniform_(emb.weight.data)
self.atom_embedding_list.append(emb)
if self.num_scalar_features > 0:
self.linear = torch.nn.Linear(self.num_scalar_features, emb_dim)
if self.lm_embedding_type is not None:
if self.lm_embedding_type == 'esm':
self.lm_embedding_dim = 1280
else: raise ValueError('LM Embedding type was not correctly determined. LM embedding type: ', self.lm_embedding_type)
self.lm_embedding_layer = torch.nn.Linear(self.lm_embedding_dim + emb_dim, emb_dim)
def forward(self, x):
x_embedding = 0
if self.lm_embedding_type is not None:
assert x.shape[1] == self.num_categorical_features + self.num_scalar_features + self.lm_embedding_dim
else:
assert x.shape[1] == self.num_categorical_features + self.num_scalar_features
for i in range(self.num_categorical_features):
x_embedding += self.atom_embedding_list[i](x[:, i].long())
if self.num_scalar_features > 0:
x_embedding += self.linear(x[:, self.num_categorical_features:self.num_categorical_features + self.num_scalar_features])
if self.lm_embedding_type is not None:
x_embedding = self.lm_embedding_layer(torch.cat([x_embedding, x[:, -self.lm_embedding_dim:]], axis=1))
return x_embedding
边表征
就是MLP映射层
nn.Sequential(nn.Linear(in_lig_edge_features + sigma_embed_dim + distance_embed_dim, ns),nn.ReLU(), nn.Dropout(dropout),nn.Linear(ns, ns))
原子距离分布表征
class GaussianSmearing(torch.nn.Module):
# used to embed the edge distances 用于嵌入边缘距离
def __init__(self, start=0.0, stop=5.0, num_gaussians=50):
super().__init__()
offset = torch.linspace(start, stop, num_gaussians)
self.coeff = -0.5 / (offset[1] - offset[0]).item() ** 2
self.register_buffer('offset', offset)
def forward(self, dist):
dist = dist.view(-1, 1) - self.offset.view(1, -1)
return torch.exp(self.coeff * torch.pow(dist, 2))
等变卷积(旋转平移不变性)
作者等变卷积使用的是e3nn框架
使用e3nn实现一个等变卷积 o3.FullyConnectedTensorProduct(in_irreps, sh_irreps, out_irreps, shared_weights=False)。
我们将执行这个公式:
f
j
⊗
(
h
(
∥
x
i
j
∥
)
)
Y
(
x
i
j
/
∥
x
i
j
∥
)
f_j \otimes\left(h\left(\left\|x_{i j}\right\|\right)\right) Y\left(x_{i j} /\left\|x_{i j}\right\|\right)
fj⊗(h(∥xij∥))Y(xij/∥xij∥)
然后归一化以及聚合:
f i ′ = 1 z ∑ j ∈ ∂ ( i ) f j ⊗ ( h ( ∥ x i j ∥ ) ) Y ( x i j / ∥ x i j ∥ ) f_i^{\prime}=\frac{1}{\sqrt{z}} \sum_{j \in \partial(i)} f_j \otimes\left(h\left(\left\|x_{i j}\right\|\right)\right) Y\left(x_{i j} /\left\|x_{i j}\right\|\right) fi′=z1j∈∂(i)∑fj⊗(h(∥xij∥))Y(xij/∥xij∥)
其中:
- f j , f i ′ f_j, f_i^{\prime} fj,fi′节点是输入和输出
- z z z节点的平均度是多少
- ∂ ( i ) \partial(i) ∂(i)是节点 i i i的邻居集合
- x i j x_{i j} xij是相对向量
- h h h是一个多层感知机
- Y Y Y是球谐波
- x ⊗ ( w ) y x \otimes(w) y x⊗(w)y是 x x x和 y y y的张量积 y y y被一些权重 w w w参数化
e3nn详细教程可以参考blog
class TensorProductConvLayer(torch.nn.Module):
def __init__(self, in_irreps, sh_irreps, out_irreps, n_edge_features, residual=True, batch_norm=True, dropout=0.0,
hidden_features=None):
super(TensorProductConvLayer, self).__init__()
self.in_irreps = in_irreps
self.out_irreps = out_irreps
self.sh_irreps = sh_irreps
self.residual = residual
if hidden_features is None:
hidden_features = n_edge_features
## 全连接向量积
self.tp = tp = o3.FullyConnectedTensorProduct(in_irreps, sh_irreps, out_irreps, shared_weights=False)
self.fc = nn.Sequential(
nn.Linear(n_edge_features, hidden_features),
nn.ReLU(),
nn.Dropout(dropout),
nn.Linear(hidden_features, tp.weight_numel)
)
self.batch_norm = BatchNorm(out_irreps) if batch_norm else None
def forward(self, node_attr, edge_index, edge_attr, edge_sh, out_nodes=None, reduce='mean'):
edge_src, edge_dst = edge_index
tp = self.tp(node_attr[edge_dst], edge_sh, self.fc(edge_attr))
out_nodes = out_nodes or node_attr.shape[0]
out = scatter(tp, edge_src, dim=0, dim_size=out_nodes, reduce=reduce)
if self.residual:
padded = F.pad(node_attr, (0, out.shape[-1] - node_attr.shape[-1]))
out = out + padded
if self.batch_norm:
out = self.batch_norm(out)
return out
质心平移和旋转(对接模型)
self.center_distance_expansion = GaussianSmearing(0.0, center_max_distance, distance_embed_dim)
self.center_edge_embedding = nn.Sequential(
nn.Linear(distance_embed_dim + sigma_embed_dim, ns),
nn.ReLU(),
nn.Dropout(dropout),
nn.Linear(ns, ns)
)
self.final_conv = TensorProductConvLayer(
in_irreps=self.lig_conv_layers[-1].out_irreps,
sh_irreps=self.sh_irreps,
out_irreps=f'2x1o + 2x1e',
n_edge_features=2 * ns,
residual=False,
dropout=dropout,
batch_norm=batch_norm
)
self.tr_final_layer = nn.Sequential(nn.Linear(1 + sigma_embed_dim, ns),nn.Dropout(dropout), nn.ReLU(), nn.Linear(ns, 1))
self.rot_final_layer = nn.Sequential(nn.Linear(1 + sigma_embed_dim, ns),nn.Dropout(dropout), nn.ReLU(), nn.Linear(ns, 1))
if not no_torsion:
# torsion angles components
self.final_edge_embedding = nn.Sequential(
nn.Linear(distance_embed_dim, ns),
nn.ReLU(),
nn.Dropout(dropout),
nn.Linear(ns, ns)
)
self.final_tp_tor = o3.FullTensorProduct(self.sh_irreps, "2e")
self.tor_bond_conv = TensorProductConvLayer(
in_irreps=self.lig_conv_layers[-1].out_irreps,
sh_irreps=self.final_tp_tor.irreps_out,
out_irreps=f'{ns}x0o + {ns}x0e',
n_edge_features=3 * ns,
residual=False,
dropout=dropout,
batch_norm=batch_norm
)
self.tor_final_layer = nn.Sequential(
nn.Linear(2 * ns, ns, bias=False),
nn.Tanh(),
nn.Dropout(dropout),
nn.Linear(ns, 1, bias=False)
)
置信度预测
置信度和亲和度预测层
self.confidence_predictor = nn.Sequential(
nn.Linear(2 * self.ns if num_conv_layers >= 3 else self.ns, ns),
nn.BatchNorm1d(ns) if not confidence_no_batchnorm else nn.Identity(),
nn.ReLU(),
nn.Dropout(confidence_dropout),
nn.Linear(ns, ns),
nn.BatchNorm1d(ns) if not confidence_no_batchnorm else nn.Identity(),
nn.ReLU(),
nn.Dropout(confidence_dropout),
nn.Linear(ns, output_confidence_dim)
)
Diffusion模型
加噪过程
作者这里选择线性加噪过程, 其他加噪过程请参考blog
def get_t_schedule(inference_steps):
return np.linspace(1, 0, inference_steps + 1)[:-1]
Time Embedding
timestep embedding作者使用了两种方法, 一个是DDPM中提到的正弦嵌入, 还有一个是高斯傅立叶嵌入
def get_timestep_embedding(embedding_type, embedding_dim, embedding_scale=10000):
if embedding_type == 'sinusoidal':
emb_func = (lambda x : sinusoidal_embedding(embedding_scale * x, embedding_dim))
elif embedding_type == 'fourier':
emb_func = GaussianFourierProjection(embedding_size=embedding_dim, scale=embedding_scale)
else:
raise NotImplemented
return emb_func
forward diffusion

,配体的构象其实本质是也就是原子在三维坐标系上的集合,因此本质上也就是数据的分布。但与图片不同的是,小分子构象的正向扩散或者说是构象变化过程是存在一定限制的,配体在本身的键长和原子间的连接方式在构象转变过程中还是会保持基本不变。作者将配体构象变化的范围称为自由度,并将这个自由度划分为了三个部分。也就是文章标题中的steps,turns以及twist,分别对应着配体构象的位置变动,构象翻转以及键的扭转。这三个维度共同构成一个子空间,并且与实际上的配体构象空间相对应。这也就使得正向扩散从直接从配体构象空间采样变成了从
R
∧
3
,
S
O
(
3
)
,
T
∧
3
\mathbb{R}^{\wedge} 3, \quad SO(3), \mathbb{T}^{\wedge} 3
R∧3,SO(3),T∧3者三个维度的采样。
def set_time(complex_graphs, t_tr, t_rot, t_tor, batchsize, all_atoms, device):
complex_graphs['ligand'].node_t = {
'tr': t_tr * torch.ones(complex_graphs['ligand'].num_nodes).to(device),
'rot': t_rot * torch.ones(complex_graphs['ligand'].num_nodes).to(device),
'tor': t_tor * torch.ones(complex_graphs['ligand'].num_nodes).to(device)}
complex_graphs['receptor'].node_t = {
'tr': t_tr * torch.ones(complex_graphs['receptor'].num_nodes).to(device),
'rot': t_rot * torch.ones(complex_graphs['receptor'].num_nodes).to(device),
'tor': t_tor * torch.ones(complex_graphs['receptor'].num_nodes).to(device)}
complex_graphs.complex_t = {'tr': t_tr * torch.ones(batchsize).to(device),
'rot': t_rot * torch.ones(batchsize).to(device),
'tor': t_tor * torch.ones(batchsize).to(device)}
if all_atoms:
complex_graphs['atom'].node_t = {
'tr': t_tr * torch.ones(complex_graphs['atom'].num_nodes).to(device),
'rot': t_rot * torch.ones(complex_graphs['atom'].num_nodes).to(device),
'tor': t_tor * torch.ones(complex_graphs['atom'].num_nodes).to(device)}
reverse diffusion
对steps,turns以及twist三个自由度进行采样
tr_sigma, rot_sigma, tor_sigma = t_to_sigma(t_tr, t_rot, t_tor)
噪声预测:
with torch.no_grad():
tr_score, rot_score, tor_score = model(complex_graph_batch)
去噪过程:
tr_g = tr_sigma * torch.sqrt(torch.tensor(2 * np.log(model_args.tr_sigma_max / model_args.tr_sigma_min)))
rot_g = 2 * rot_sigma * torch.sqrt(torch.tensor(np.log(model_args.rot_sigma_max / model_args.rot_sigma_min)))
if ode:
tr_perturb = (0.5 * tr_g ** 2 * dt_tr * tr_score.cpu()).cpu()
rot_perturb = (0.5 * rot_score.cpu() * dt_rot * rot_g ** 2).cpu()
else:
tr_z = torch.zeros((b, 3)) if no_random or (no_final_step_noise and t_idx == inference_steps - 1) \
else torch.normal(mean=0, std=1, size=(b, 3))
tr_perturb = (tr_g ** 2 * dt_tr * tr_score.cpu() + tr_g * np.sqrt(dt_tr) * tr_z).cpu()
rot_z = torch.zeros((b, 3)) if no_random or (no_final_step_noise and t_idx == inference_steps - 1) \
else torch.normal(mean=0, std=1, size=(b, 3))
rot_perturb = (rot_score.cpu() * dt_rot * rot_g ** 2 + rot_g * np.sqrt(dt_rot) * rot_z).cpu()
if not model_args.no_torsion:
tor_g = tor_sigma * torch.sqrt(torch.tensor(2 * np.log(model_args.tor_sigma_max / model_args.tor_sigma_min)))
if ode:
tor_perturb = (0.5 * tor_g ** 2 * dt_tor * tor_score.cpu()).numpy()
else:
tor_z = torch.zeros(tor_score.shape) if no_random or (no_final_step_noise and t_idx == inference_steps - 1) \
else torch.normal(mean=0, std=1, size=tor_score.shape)
tor_perturb = (tor_g ** 2 * dt_tor * tor_score.cpu() + tor_g * np.sqrt(dt_tor) * tor_z).numpy()
torsions_per_molecule = tor_perturb.shape[0] // b
else:
tor_perturb = None