参考:https://zhuanlan.zhihu.com/p/537206999
https://scanpy.readthedocs.io/en/stable/api.html
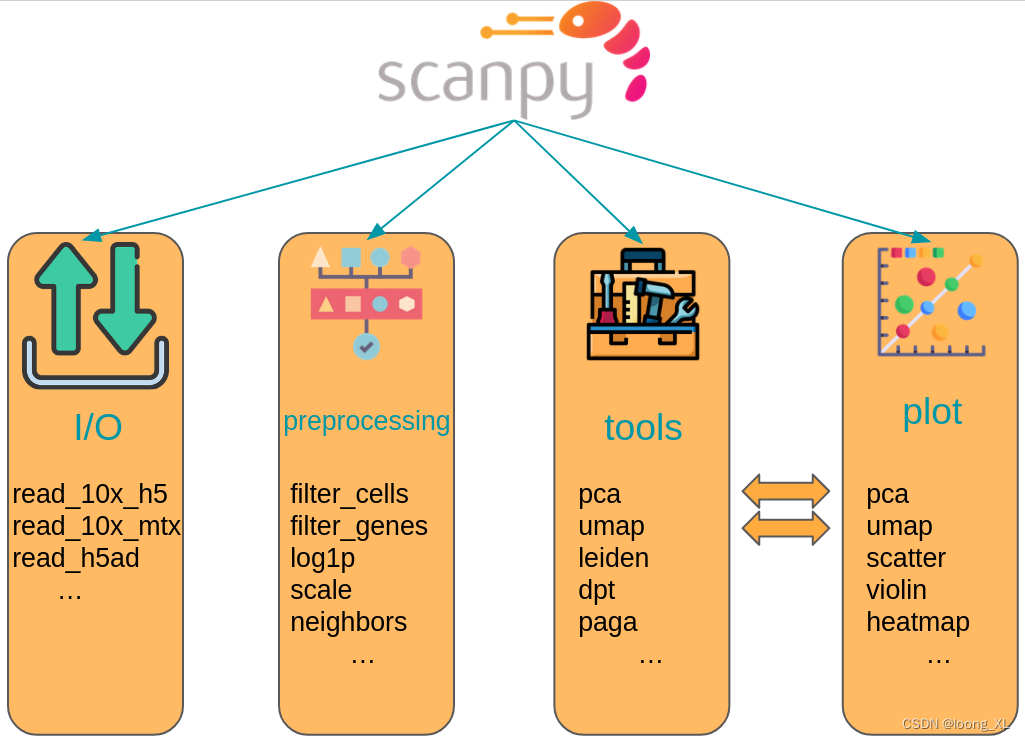
scanpy python包主要分四个模块:
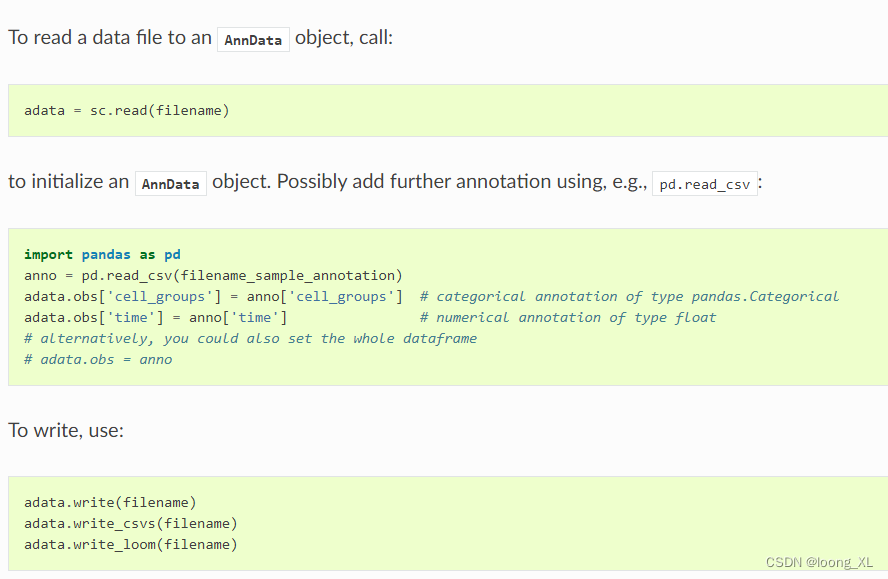
1)read 读写模块、
https://scanpy.readthedocs.io/en/stable/api.html#reading
2)pp Preprocessing 预处理模块
https://scanpy.readthedocs.io/en/stable/api.html#module-scanpy.pp

3)tl Tools工具箱模块,包括降维聚类等算法
https://scanpy.readthedocs.io/en/stable/api.html#module-scanpy.tl

3)pl Plotting画图模块
https://scanpy.readthedocs.io/en/stable/api.html#module-scanpy.pl

使用案例
import numpy as np
import pandas as pd
import scanpy as sc
## 加载数据
adata = sc.datasets.pbmc3k()
adata.var_names_make_unique()
#质量控制
# 查看高表达的前20个基因
sc.pl.highest_expr_genes(adata, n_top=20, save='_pbmc3k.png')
#######预处理##############
sc.pp.filter_cells(adata, min_genes=200)
sc.pp.filter_genes(adata, min_cells=3)
adata.var['mt'] = adata.var_names.str.startswith('MT-') # annotate the group of mitochondrial genes as 'mt'
sc.pp.calculate_qc_metrics(adata, qc_vars=['mt'], percent_top=None, log1p=False, inplace=True)
#查看
sc.pl.violin(adata, ['n_genes_by_counts', 'total_counts', 'pct_counts_mt'],
jitter=0.4, multi_panel=True, save='_pbmc3k.png')
adata = adata[adata.obs.n_genes_by_counts < 2500, :]
adata = adata[adata.obs.pct_counts_mt < 5, :]
#总计数归一化、对数化
sc.pp.normalize_total(adata, target_sum=1e4)
sc.pp.log1p(adata)
# 识别高度可变的基因
sc.pp.highly_variable_genes(adata, min_mean=0.0125, max_mean=3, min_disp=0.5)
sc.pl.highly_variable_genes(adata, save='_pbmc3k.png')
# 保存原始数据
adata.raw = adata
# 过滤
adata = adata[:, adata.var.highly_variable]
# 将数据缩放到单位方差
sc.pp.regress_out(adata, ['total_counts', 'pct_counts_mt'])
sc.pp.scale(adata, max_value=10)
#######降维聚类分析##############
sc.tl.pca(adata, svd_solver='arpack')
sc.pl.pca(adata, color='CST3', save='_pbmc3k_CST3.png')
sc.pl.pca_variance_ratio(adata, log=True, save='_pbmc3k_pc.png')
sc.pp.neighbors(adata, n_neighbors=10, n_pcs=40)
sc.tl.umap(adata)
sc.pl.umap(adata, color=['CST3', 'NKG7', 'PPBP'], save='_pbmc3k_CST3_NKG7_PPBP.png')
sc.tl.leiden(adata)
sc.pl.umap(adata, color=['leiden'], save='_pbmc3k_leiden.png')