Title
题目
SAMUS: Adapting Segment Anything Model for Clinically-Friendly and Generalizable Ultrasound Image Segmentation
SAMUS:适应临床友好型和泛化的超声图像分割的Segment Anything模型
01
文献速递介绍
医学图像分割是一项关键技术,用于辨识和突出显示医学图像中的特定器官、组织和病变,是计算机辅助诊断系统的一个组成部分(刘等,2021)。为自动医学图像分割提出了众多深度学习模型,展示了巨大的潜力(Ronneberger, Fischer, 和 Brox,2015;吴等,2022)。然而,这些模型是为特定对象量身定做的,并且在应用于其他对象时需要重新训练,给临床使用带来了极大的不便。
段落任何模型(SAM),作为一个多用途的视觉分割基础模型,由于其在多样对象上的显著分割能力和强大的零次学习泛化能力,获得了相当的赞誉(Kirillov等,2023)。根据用户提示,包括点、边界框和粗略遮罩,SAM能够对应分割对象。因此,通过简单的提示,SAM可以轻松适应各种分割应用。这一范式使得将多个单一医学图像分割任务整合进一个统一框架(即,一个通用模型)成为可能,大大促进了临床部署(黄等,2023)。
尽管构建了迄今为止最大的数据集(即,SA-1B),SAM在医学领域遭遇快速性能下降,原因是可靠的临床注释稀缺(黄等,2023)。一些基础模型已经被提出,通过在医学数据集上调整SAM来适应医学图像分割(马和王,2023;吴等,2023)。然而,与SAM相同,它们在特征建模之前对输入图像进行16×的无重叠分割,这破坏了识别小目标和边界所需的局部信息,使它们难以分割具有复杂/线状形状、弱边界、小尺寸或低对比度的临床对象。此外,它们中的大多数要求输入尺寸为1024×1024,由于生成的长输入序列,给GPU消耗带来了巨大负担。
在本文中,我们介绍SAMUS,将SAM的卓越分割性能和强大的泛化能力转移到医学图像分割领域,同时降低了计算复杂度。SAMUS继承了SAM的ViT图像编码器、提示编码器和掩码解码器,对图像编码器进行了量身定制的设计。首先,我们通过减小输入尺寸来缩短ViT分支的序列长度,以降低计算复杂度。然后,开发了特征适配器和位置适配器,以从自然域微调ViT图像编码器到医学域。为了补充ViT图像编码器中的局部(即,低级)信息,我们引入了一个并行的CNN分支图像编码器,与ViT分支并行运行,并提出了一个跨分支注意力模块,使ViT分支中的每个补丁都能从CNN分支吸收局部信息。
Abstract-Background
摘要
Segment anything model (SAM), an eminent universal im age segmentation model, has recently gathered considerable attention within the domain of medical image segmenta tion. Despite the remarkable performance of SAM on natu ral images, it grapples with significant performance degrada tion and limited generalization when confronted with med ical images, particularly with those involving objects of low contrast, faint boundaries, intricate shapes, and diminu tive sizes. In this paper, we propose SAMUS, a universal model tailored for ultrasound image segmentation. In con trast to previous SAM-based universal models, SAMUS pur sues not only better generalization but also lower deploy ment cost, rendering it more suitable for clinical applications. Specifically, based on SAM, a parallel CNN branch is intro duced to inject local features into the ViT encoder through cross-branch attention for better medical image segmenta tion. Then, a position adapter and a feature adapter are de veloped to adapt SAM from natural to medical domains and from requiring large-size inputs (1024×1024) to small-size inputs (256×256) for more clinical-friendly deployment. A comprehensive ultrasound dataset, comprising about 30k im ages and 69k masks and covering six object categories, is collected for verification. Extensive comparison experiments demonstrate SAMUS’s superiority against the state-of-the-art task-specific models and universal foundation models under both task-specific evaluation and generalization evaluation. Moreover, SAMUS is deployable on entry-level GPUs, as it has been liberated from the constraints of long sequence encoding. The code, data, and models will be released at https://github.com/xianlin7/SAMUS.
段落任何模型(SAM),作为一个杰出的通用图像分割模型,最近在医疗图像分割领域内获得了相当多的关注。尽管SAM在自然图像上的性能表现显著,但面对医学图像,尤其是那些涉及低对比度、界限模糊、形状复杂和尺寸微小的对象时,它却面临着显著的性能下降和有限的泛化能力。在本文中,我们提出了SAMUS,一个专为超声图像分割定制的通用模型。与之前基于SAM的通用模型相比,SAMUS不仅追求更好的泛化能力,也追求更低的部署成本,使其更适合临床应用。
具体而言,基于SAM,通过引入一个并行的CNN分支,通过跨分支注意力将局部特征注入ViT编码器,以实现更好的医学图像分割。然后,开发了位置适配器和特征适配器,以将SAM从自然领域适配到医学领域,并从需要大尺寸输入(1024×1024)到小尺寸输入(256×256)进行适配,以实现更加临床友好的部署。收集了一个全面的超声数据集,包括大约30k图像和69k掩模,覆盖六个对象类别,用于验证。广泛的比较实验表明,SAMUS在任务特定评估和泛化评估下,都优于最先进的任务特定模型和通用基础模型。
此外,SAMUS可以部署在入门级GPU上,因为它已经摆脱了长序列编码的限制。代码、数据和模型将在https://github.com/xianlin7/SAMUS发布
Conclusions
结论
In this paper, we propose SAMUS, a universal founda tion model derived from SAM, for clinically-friendly and generalizable ultrasound image segmentation. Specifically, we present a parallel CNN branch image encoder, a fea ture adapter, a position adapter, and a cross-branch atten tion module to enrich the features for small-size objects and boundary areas while reducing GPU consumption. Further more, we construct a large ultrasound image dataset US30K, consisting of 30,106 images and 68,570 masks for eval uation and potential clinical usage. Experiments on both seeable and unseen domains demonstrate the outstanding segmentation ability and strong generalization ability of SAMUS. Moreover, the GPU memory cost of SAMUS is merely 28% of that required to train the entire SAM, and SAMUS is about 3× faster than SAM for inference.
在本文中,我们提出了SAMUS,一个基于SAM衍生的通用基础模型,用于临床友好和可泛化的超声图像分割。具体而言,我们展示了一个并行的CNN分支图像编码器、一个特征适配器、一个位置适配器和一个跨分支注意力模块,以丰富小尺寸对象和边界区域的特征,同时减少GPU消耗。此外,我们构建了一个大型超声图像数据集US30K,包含30,106张图像和68,570个掩模,用于评估和潜在的临床使用。在可见和未见领域的实验展示了SAMUS的卓越分割能力和强大的泛化能力。此外,SAMUS的GPU内存成本仅为训练整个SAM所需的28%,且SAMUS的推理速度大约是SAM的3倍。
Method
方法
As depicted in Fig. 8, the overall architecture of SAMUS is inherited from SAM, retaining the structure and param eters of the prompt encoder and the mask decoder without any adjustment. Comparatively, the image encoder is care fully modified to address the challenges of inadequate local features and excessive computational memory consumption, making it more suitable for clinically-friendly segmentation. Major modifications include reducing the input size, over
lapping the patch embedding, introducing adapters to the ViT branch, adding a CNN branch, and introducing cross branch attention (CBA). Specifically, the input spatial reso ution is scaled down from 1024 × 1024 pixels to 256 × 256 pixels, resulting in a substantial reduction in GPU memory cost due to the shorter input sequence in transformers. The overlapped patch embedding uses the same parameters as the patch embedding in SAM while its patch stride is half to the original stride, well keeping the information from patch boundaries. Adapters in the ViT branch include a position adapter and five feature adapters. The position adapter is to accommodate the global position embedding in shorter sequences due to the smaller input size. The first feature adapter follows the overlapped patch embedding to align in put features with the required feature distribution of the pre trained ViT image encoder. The remaining feature adapters are attached to the residual connections of the feed-forward network in the global transformer to fine-tune the pre-trained image encoder. In terms of the CNN branch, it is parallel to the ViT branch, providing complementary local information to the latter through the CBA module, which takes the ViT branch features as the query and builds global dependency with features from the CNN branch. It should be noted that CBA is only integrated into each global transformer. Finallythe outputs of both the two branches are combined as the fi nal image feature embedding of SAMUS.
如图8所示,SAMUS的总体架构继承自SAM,保留了提示编码器和掩码解码器的结构和参数,没有任何调整。相比之下,图像编码器经过仔细修改,以解决不足的局部特征和过多的计算内存消耗问题,使其更适合临床友好的分割。主要修改包括减小输入尺寸、重叠的补丁嵌入、向ViT分支引入适配器、增加CNN分支,并引入跨分支注意力(CBA)。具体而言,输入的空间分辨率从1024×1024像素降低到256×256像素,由于变换器中输入序列的缩短,显著降低了GPU内存成本。重叠的补丁嵌入使用与SAM中的补丁嵌入相同的参数,而其补丁步长是原始步长的一半,很好地保留了补丁边界的信息。ViT分支中的适配器包括一个位置适配器和五个特征适配器。位置适配器是为了适应更小输入尺寸下的短序列中的全局位置嵌入。第一个特征适配器跟随重叠的补丁嵌入,以将输入特征与预训练ViT图像编码器所需的特征分布对齐。其余特征适配器附加在全局变换器的前馈网络的残差连接上,以微调预训练的图像编码器。就CNN分支而言,它与ViT分支平行,通过CBA模块提供补充的局部信息给后者,CBA模块将ViT分支特征作为查询,并与CNN分支的特征建立全局依赖关系。应该注意的是,CBA仅集成在每个全局变换器中。最后,两个分支的输出合并为SAMUS的最终图像特征嵌入。
Figure
图
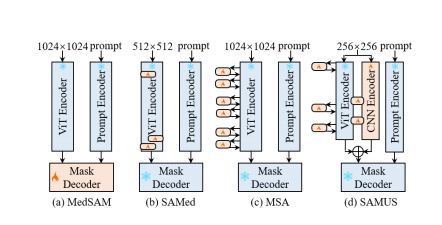
Figure 1: Structure comparison of different SAM-based foundation models for medical image segmentation.
图1:基于SAM的不同医学图像分割基础模型的结构比较。
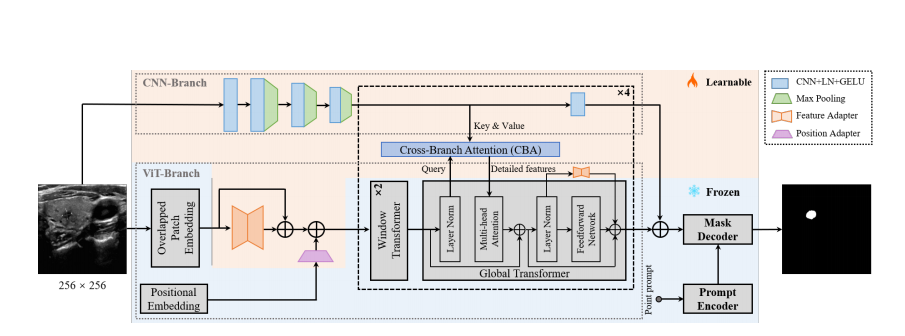
Figure 2: Overview of the proposed SAMUS.
图2:提出的SAMUS的概述。
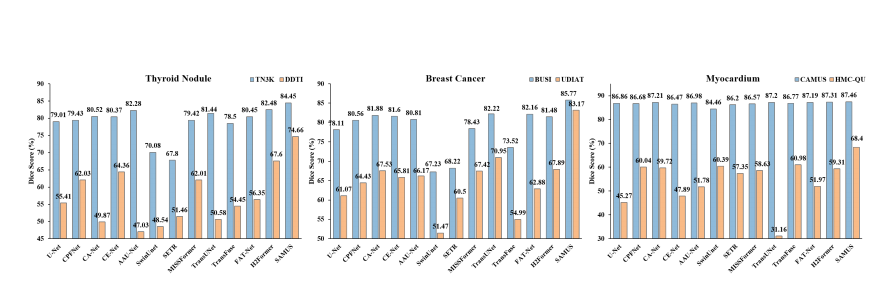
Figure 3: Comparison between SAMUS and task-specific methods evaluated on seeable (marked in blue) and unseen datasets(marked in orange).
图3:在可见(用蓝色标记)和未见(用橙色标记)数据集上评估的SAMUS与任务特定方法的比较。
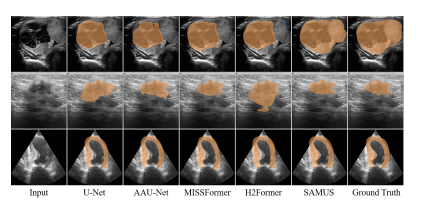
Figure 4: Qualitative comparisons between SAMUS and task-specific methods. From top to bottom are examples of segmenting thyroid nodule, breast cancer, and myocardium.
图4:SAMUS与任务特定方法之间的定性比较。从上到下分别是分割甲状腺结节、乳腺癌和心肌的示例。

Figure 5: Qualitative comparisons between SAMUS and foundation models. From top to bottom are examples of seg menting thyroid nodule, breast cancer, and myocardium.
图5:SAMUS与基础模型之间的定性比较。从上到下分别是分割甲状腺结节、乳腺癌和心肌的示例。
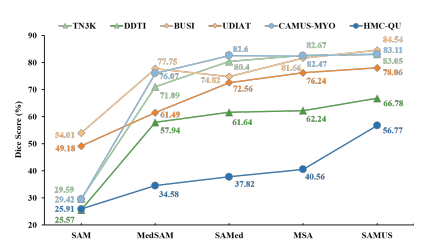
Figure 6: Segmentation and generalization ability compari son of our SAMUS and other foundation models on seeable (in light color) and unseen (in dark color) US30K data.
图6:我们的SAMUS与其他基础模型在可见的(浅色)和未见的(深色)US30K数据上的分割和泛化能力比较。
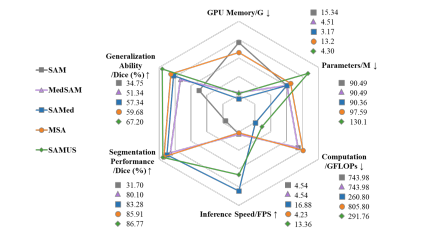
Figure 7: Comparison of SAMUS and foundation models on GPU memory cost, model parameters, computational com plexity, inference speed, performance, and generalization.
图7:SAMUS与基础模型在GPU内存成本、模型参数、计算复杂度、推理速度、性能和泛化能力上的比较。
Table
表
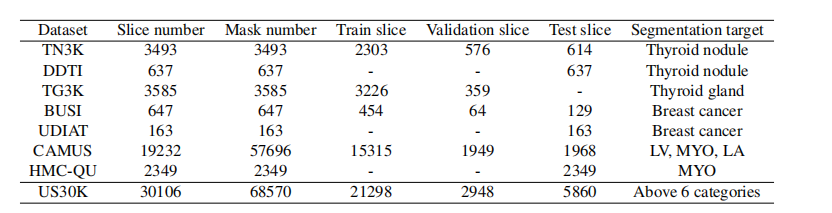
Table 1: Summary of the datasets in US30K. LV, MYO, and LA are short for the left ventricle, myocardium, and left atrium.
表1:US30K数据集摘要。LV、MYO和LA分别代表左心室、心肌和左心房。
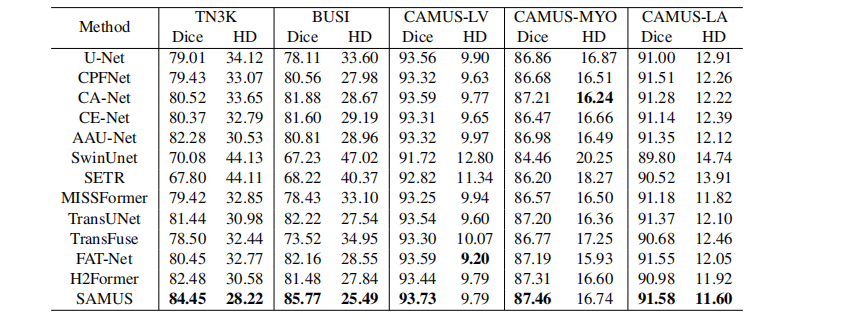
Table 2: Quantitative comparison of our SAMUS and SOTA task- specific methods on segmenting thyroid nodule (TN3K), breast cancer (BUSI), left ventricle (CAMUS-LV), myocardium (CAMUS-MYO), and left atrium (CAMUS- LA). The perfor mance is evaluated by the Dice score (%) and Hausdorff distance (HD). The best results are marked in bold.
表2:我们的SAMUS与SOTA任务特定方法在分割甲状腺结节(TN3K)、乳腺癌(BUSI)、左心室(CAMUS-LV)、心肌(CAMUS-MYO)和左心房(CAMUS-LA)上的定量比较。性能通过Dice得分(%)和Hausdorff距离(HD)评估。最佳结果以粗体标记。
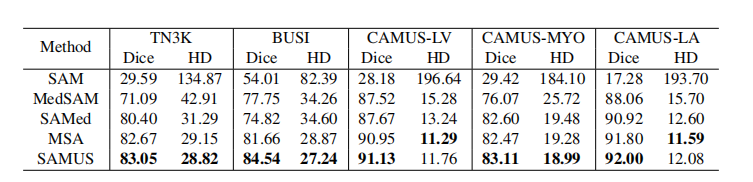
Table 3: Quantitative comparison of our SAMUS and other foundation models on seeable US30K data. The performance is evaluated by the Dice score (%) and Hausdorff distance (HD).
表3:我们的SAMUS与其他基础模型在可见的US30K数据上的定量比较。性能通过Dice得分(%)和Hausdorff距离(HD)评估。

Table 4: Ablation study on different component combinations of SAMUS on the thyroid nodule and breast cancer segmentation. F-Adapter and P-Adapter represent the feature adapter and the position adapter respectively
表4:SAMUS在甲状腺结节和乳腺癌分割上不同组件组合的消融研究。F-Adapter和P-Adapter分别代表特征适配器和位置适配器。
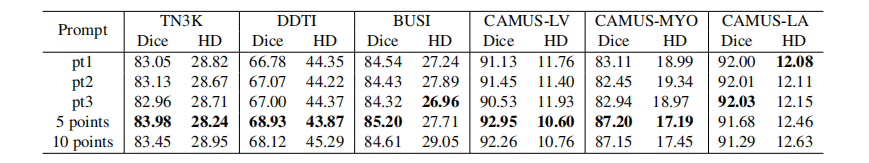
Table 5: Ablation study of different prompts. Pt1, pt2, and pt3 represent the single-point prompt in different (randomly deter mined) foreground positions. Multipoint prompts are generated by random sampling on the foreground areas.
表5:不同提示的消融研究。Pt1、pt2和pt3代表在不同(随机确定的)前景位置的单点提示。多点提示通过在前景区域的随机抽样生成。