算法原理
卷积神经网络(Convolutional Neural Networks,CNN)是深度学习中最常见的一种
算法,它具有强大的特征学习能力。CNN 通过结合局部感知区域、共享权重、空间或者
时间上的降采样来充分利用数据本身包含的局部性等特征,优化网络结构,并且保证一
定程度上的位移和变形的不变性。因此,CNN 被广泛应用在图像分类,语音识别,目标
检测和人脸识别等领域。一般而言,一个简单的卷积神经网络结构通常由若干个卷积层,
池化层和全连接层组成,如图 1 所示。
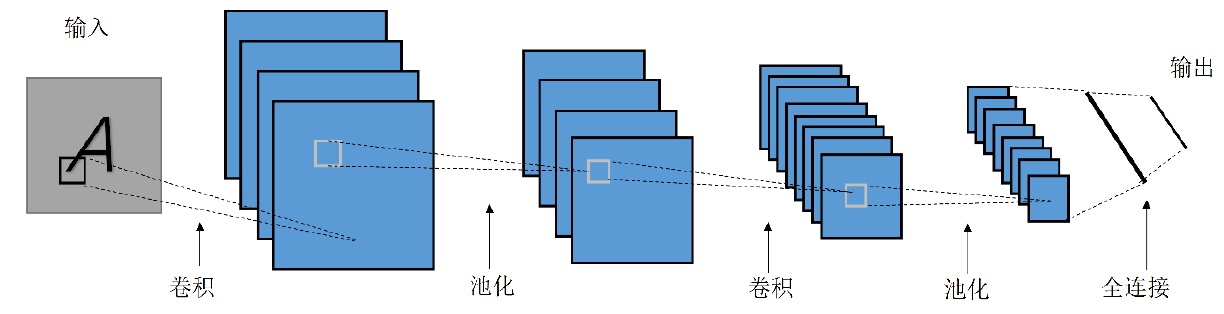
图1 基本的卷积神经网络结构
(1)卷积层。卷积层是一个特征学习的过程,其核心是利用卷积核在输入的图像中
上下滑动,图像上的像素值与卷积核内的值做卷积操作。卷积层的功能是对输入数据进
行特征提取,其内部包含多个卷积核,组成卷积核的每个元素都对应一个权重系数和一
个偏置值。卷积层内每个神经元都与前一层中位置接近的区域的多个神经元相连,区域
的大小取决于卷积核的大小。卷积是一种特殊的线性操作,卷积核会有规律地扫过输入
图像,在感受野内对输入特征做矩阵元素乘法求和并叠加偏值量,由公式(2-1)所示。

(2)池化层。也被称为下采样层,通常用于卷积层之后。池化层包含预设定的池化
函数,其功能是将特征图中单个点的值替换为其相邻区域的特征图统计量。池化层选取
池化区域与卷积核扫描特征图步骤相同,由池化大小(Size)、步长(Stride)和填充
(Padding)控制。池化的目的是用来缩小卷积后的特征图尺寸,池化后的特征图保留了
原始特征图的轮廓信息,以提高计算速度,加强特征的鲁棒性。
(3)全连接层。将前层学习到的局部特征进行重组,得到全局特征信息。全连接层
的输出向量维度(CN× 1)与分类个数(N)一致,全连接层的输出值通过输出层的分类
器映射为相对概率,根据概率值判断并输出最终所属结果。常用的分类器有逻辑回归函
数(Softmax Function)、支持向量机(Support Vector Machine, SVM)等。
为了强化网络的特征的学习能力,通常在卷积层后加入非线性激活函数,用于提升
特征信息的非线性化。常见的激活函数有 Sigmoid 函数,Tanh 函数和 ReLU(Rectified
Linear Unit, ReLU)函数]等。相比 Sigmoid 和 Tanh,ReLU 函数更简单,它具有更快
的收敛速度,在一定程度上避免了梯度爆炸和梯度消失问题。计算如式(2-2)所示。

其中,x 为输入特征张量。
数据集

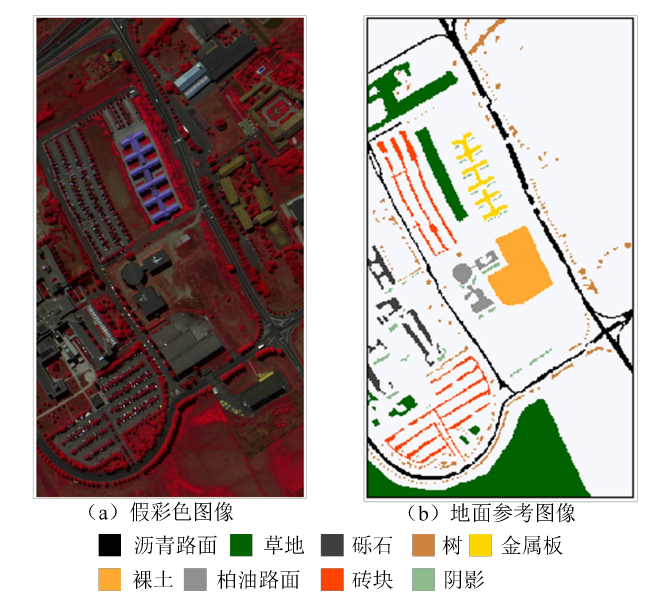
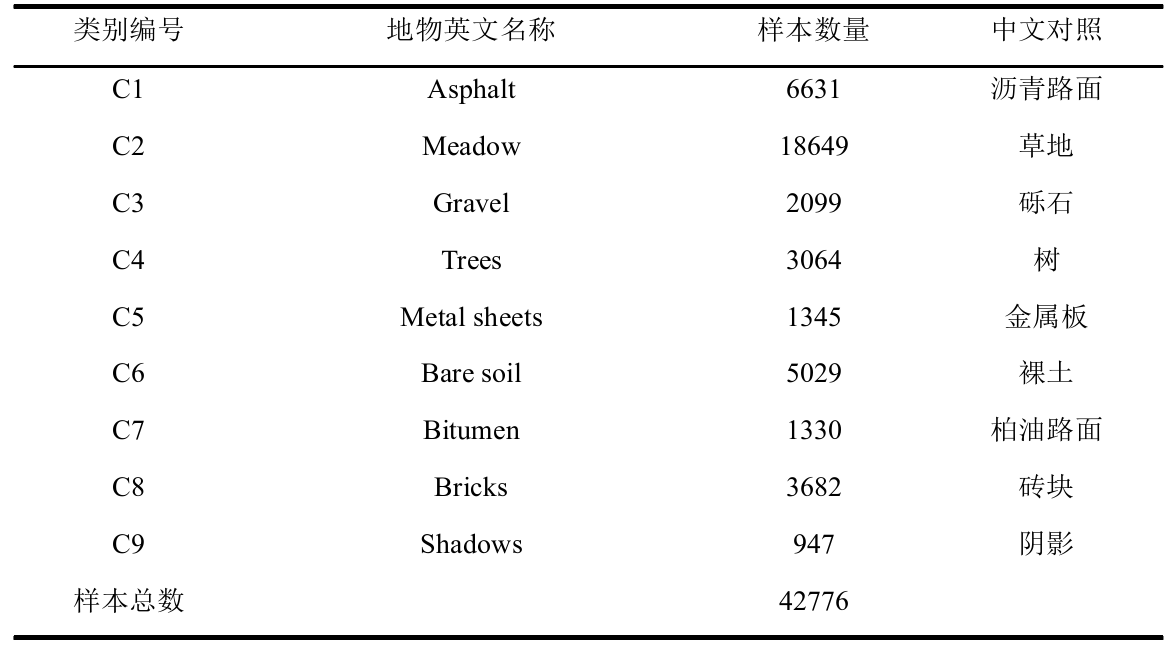
Pavia University 数据集来源于 ROSIS 传感器,反映了意大利北部地区帕维
亚大学校园及其周边场景,影像大小 610 像素×340 像素,波长范围 0.43-8.6 um,
空间分辨率 1.3m,去除噪声波段后剩余 103 个波段用于分类。该数据集共有 9
类地物,42776 个样本,详细类别信息见表 2.2。该区域的假彩色图像和地面参
考图像如图 2 所示。其中图左为该区域的假彩色图(合成波段:红光
波段 90、绿光波段 60 和蓝光波段 30),图 右为该区域的地面参考图像。
KSC数据由 AVIRIS 传感器在佛罗里达州肯尼迪太空中心于1996年3月23日拍摄。这个数据包含了224个波段,经过水汽噪声去除后还剩下176个波段,空间分辨率是18米,一共有13个类别。
下面链接有很多数据集 该程序适用于很多高光谱数据集
https://blog.csdn.net/weixin_39860349/article/details/111263380
程序运行环境: python 3.6 TensorFlow 1.15 keras 2.3.1
1D-CNN
注:没怎么调参,可根据实际情况进行优化
#1DCNN光谱特征分类
import keras
from keras.models import Sequential
from keras.layers import Flatten,Dense,Conv1D,MaxPooling1D,BatchNormalization,Dropout
from sklearn.model_selection import train_test_split
from keras.optimizers import Adam
from keras.losses import categorical_crossentropy
import matplotlib.pyplot as plt
import fun
#PU
path='PaviaU.mat'
name='paviaU'
path_gt='PaviaU_gt.mat'
name_gt='paviaU_gt'
shape=(103,1) # 根据实际波段进行修改
save_path="PaviaU_1DCNN.h5"
classes=10 # 根据实际波段进行修改
#KSC
# path='KSC.mat'
# name='KSC'
# path_gt='KSC_gt.mat'
# name_gt='KSC_gt'
# shape=(176,1)
# save_path="KSC_1DCNN.h5"
# classes=14
model = Sequential()
model.add(Conv1D(32, kernel_size=1, activation='relu', input_shape=shape))
model.add(BatchNormalization())
model.add(Conv1D(48, kernel_size=1, activation='relu'))
model.add(BatchNormalization())
model.add(Conv1D(120, kernel_size=1, activation='relu'))
model.add(BatchNormalization())
model.add(MaxPooling1D(pool_size=1))
model.add(Flatten())
model.add(Dense(128, activation='relu'))
model.add(Dense(64, activation='relu'))
model.add(Dropout(0.25))
model.add(Dense(classes, activation='softmax'))
model.compile(loss=categorical_crossentropy,
optimizer=keras.optimizers.Adam(lr=0.001),
metrics=['accuracy'])
data_D,data_L=fun.data_get_1D(path,name,path_gt,name_gt)
train_D,test_D,train_L,test_L=train_test_split(data_D,data_L,test_size=0.8)
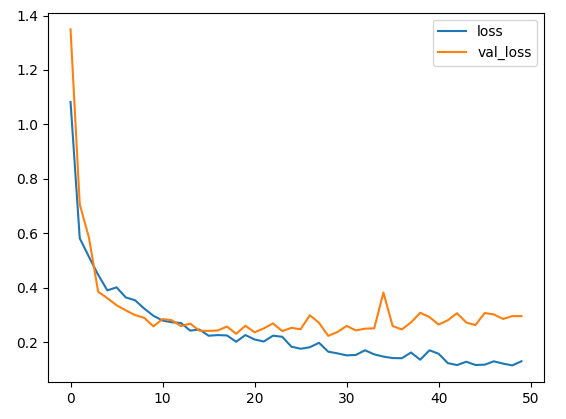
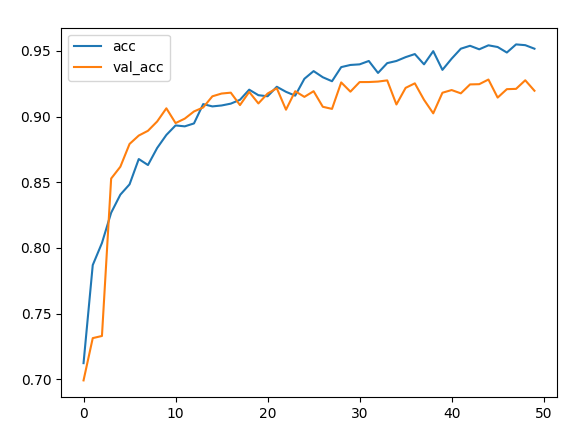
histoty=model.fit(train_D,train_L,epochs=50,batch_size=64,validation_data=(test_D ,test_L))
model.save(save_path)
plt.plot(histoty.history['loss'],label='loss')
plt.plot(histoty.history['val_loss'],label='val_loss')
plt.legend()
plt.show()
plt.plot(histoty.history['accuracy'],label='acc')
plt.plot(histoty.history['val_accuracy'],label='val_acc')
plt.legend()
plt.show()
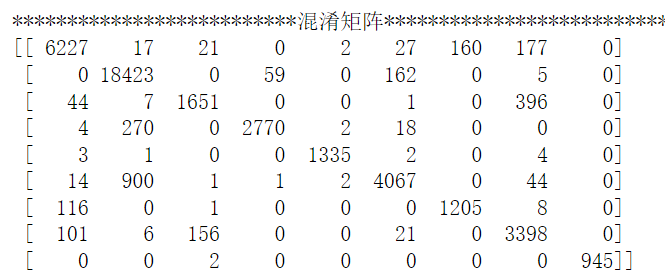
con, acc, pred_D=fun.pred(model,data_D,data_L)
print('**********混淆矩阵**********')
print(con)
fun.show(path_gt, name_gt)
fun.pred_show(path_gt,name_gt,pred_D,100)
子函数 fun.py
from scipy.io import loadmat
from keras.utils import to_categorical
import numpy as np
from sklearn import preprocessing
from sklearn.metrics import accuracy_score,confusion_matrix
from functools import reduce
import spectral
import cv2
def data_get_1D(path,name,path_gt,name_gt):
input_image = loadmat(path)[name]
output_image = loadmat(path_gt)[name_gt]
data_list = []
label_list = []
for i in range(output_image.shape[0]):
for j in range(output_image.shape[1]):
if output_image[i][j] != 0:
data_list.append(input_image[i][j])
label_list.append(output_image[i][j])
data_D = np.array([data_list])
data_L = np.array(label_list)
data_L = to_categorical(data_L)
data_D=data_D.reshape([data_D.shape[1],data_D.shape[2]])
data_D= preprocessing.StandardScaler().fit_transform(data_D)
data_D = data_D.reshape([data_D.shape[0], data_D.shape[1], 1])
return data_D,data_L
def standardScaler(X):
means = np.mean(X, axis=(0, 2)) # 计算均值,先计算第0维度的,后计算第2维度的,得到一个shape为3,的矩阵
stds = np.std(X, axis=(0, 2)) # 计算方差,得到一个shape为3,的矩阵
means = means.reshape(1, -1, 1) # 转换为和X相同维度的矩阵,此时reshape后的矩阵维度为(1,3,1)
stds = stds.reshape(1, -1, 1) # 同上
X = (X - means) / stds
return X
def data_get(path,name,path_gt,name_gt,len):
a=int((len-1)/2)
input_image = loadmat(path)[name]
output_image = loadmat(path_gt)[name_gt]
#边缘填充
input_image=cv2.copyMakeBorder(input_image, a, a, a, a,borderType=cv2.BORDER_REFLECT_101)
output_image=cv2.copyMakeBorder(output_image, a, a, a, a,borderType=cv2.BORDER_REFLECT_101)
data_list = []
label_list = []
for i in range(a, output_image.shape[0] - a):
for j in range(a, output_image.shape[1] - a):
if output_image[i][j] != 0:
data_list.append((np.float32(standardScaler(input_image[i - a:i + a+1, j - a:j + a+1, :]))))
label_list.append(output_image[i][j])
data_D = np.array([data_list])
data_L = np.array(label_list)
data_L = to_categorical(data_L)
data_2D = data_D.reshape([data_D.shape[1], len, len, data_D.shape[4]])
data_3D = data_D.reshape([data_D.shape[1],len, len, data_D.shape[4],1])
return data_2D,data_3D,data_L
def pred(model,data_D,data_L):
pred_D = np.argmax(model.predict(data_D), axis=1)
con = confusion_matrix(np.argmax(data_L, axis=1), pred_D)
acc = accuracy_score(np.argmax(data_L, axis=1), pred_D)
return con,acc,pred_D
def number(path_gt,name_gt):
output_image = loadmat(path_gt)[name_gt]
dict_k = {}
for i in range(output_image.shape[0]): # 行
for j in range(output_image.shape[1]): # 列
# if output_image[i][j] in [m for m in range(1,17)]:
if output_image[i][j] in [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13]:
if output_image[i][j] not in dict_k:
dict_k[output_image[i][j]] = 0
dict_k[output_image[i][j]] += 1
s=reduce(lambda x, y: x + y, dict_k.values())
return dict_k,s
def show(path_gt,name_gt):
output_image = loadmat(path_gt)[name_gt]
return spectral.imshow(classes=output_image.astype(int), figsize=(9, 9))
def pred_show(path_gt,name_gt,predict_label,len):
a=int((len-1)/2)
output_image = loadmat(path_gt)[name_gt]
output_image=cv2.copyMakeBorder(output_image, a, a, a, a,borderType=cv2.BORDER_CONSTANT, value=0)
new_show = np.zeros((output_image.shape[0], output_image.shape[1]))
k = 0
for i in range(a,output_image.shape[0]-a):
for j in range(a,output_image.shape[1]-a):
if output_image[i][j] != 0:
new_show[i][j] = predict_label[k]
k += 1
return spectral.imshow(classes=new_show.astype(int), figsize=(9, 9))
1D-CNN 结果
真实值:

预测值:
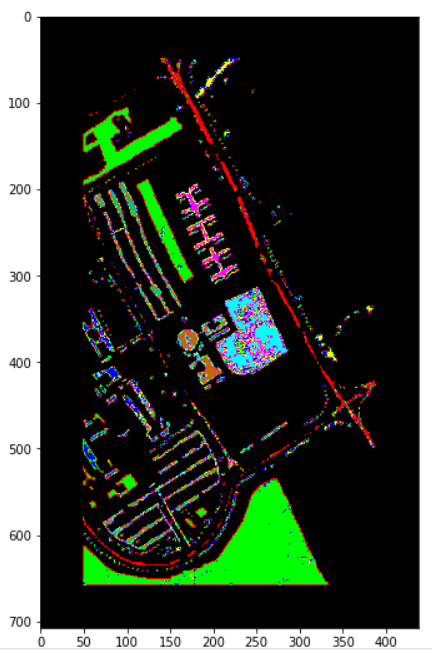
2D-CNN
import keras
from keras.layers import Flatten, Dense, BatchNormalization, Dropout, Conv2D, MaxPooling2D
from keras.models import Sequential
from sklearn.model_selection import train_test_split
from keras.optimizers import Adam
from keras.losses import categorical_crossentropy
from matplotlib import pyplot as plt
import fun
# PU
path='PaviaU.mat'
name='paviaU'
path_gt='PaviaU_gt.mat'
name_gt='paviaU_gt'
shape=(11,11,103)
classes=10
save_path="paviaU_2DCNN.h5"
#KSC
# path='data/KSC.mat'
# name='KSC'
# path_gt='data/KSC_gt.mat'
# name_gt='KSC_gt'
# shape=(11,11,176)
# save_path="KSC_2DCNN.h5"
# classes=14
model = Sequential()
model.add(Conv2D(32, kernel_size=(2, 2), activation='relu', input_shape=shape))
model.add(BatchNormalization())
model.add(Conv2D(48, kernel_size=(2, 2), activation='relu'))
model.add(BatchNormalization())
model.add(Conv2D(120, kernel_size=(2, 2), activation='relu'))
model.add(BatchNormalization())
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(128, activation='relu'))
model.add(Dropout(0.25))
model.add(Dense(64, activation='relu'))
model.add(Dropout(0.4))
model.add(Dense(classes, activation='softmax'))
model.compile(loss=categorical_crossentropy,
optimizer=keras.optimizers.Adam(lr=0.001),
metrics=['accuracy'])
data_D,data_3D,data_L=fun.data_get(path,name,path_gt,name_gt,11)
train_D,test_D,train_L,test_L=train_test_split(data_D,data_L,test_size=0.8,random_state=1)
histoty=model.fit(train_D,train_L,epochs=200,batch_size=64,validation_data=(test_D ,test_L))
model.save(save_path)
3D-CNN
from keras.layers import Conv3D,MaxPooling3D,Flatten,Dropout,Dense
from keras.models import Sequential
from sklearn.model_selection import train_test_split
from keras.optimizers import Adam
from keras.losses import categorical_crossentropy
from matplotlib import pyplot as plt
import fun
#PU
path='PaviaU.mat'
name='paviaU'
path_gt='PaviaU_gt.mat'
name_gt='paviaU_gt'
shape=(11,11,103,1)
#KSC
# path='data/KSC.mat'
# name='KSC'
# path_gt='data/KSC_gt.mat'
# name_gt='KSC_gt'
#shape=(11,11,176,1)
save_path="PU_3DCNN.h5"
classes=10
model=Sequential([
Conv3D(6,(5,5,5),strides=(1,1,1),input_shape=shape,padding="same",activation='relu'),
MaxPooling3D((2,2,2),padding="valid"),
Conv3D(12,(3,3,3),strides=(1,1,3),padding="valid",activation="relu"),
Conv3D(24,(3,1,3),strides=(1,1,3),padding="valid",activation="relu"),
Conv3D(48,(1,3,3),strides=(1,1,3),padding="valid",activation="relu"),
Flatten(),
Dropout(0.5),
Dense(96,activation='relu'),
Dense(classes,activation="softmax")
])
data_2D,data_D,data_L=fun.data_get(path,name,path_gt,name_gt,11)
train_D,test_D,train_L,test_L=train_test_split(data_D,data_L,test_size=0.9)
model.compile(optimizer=Adam(lr=0.001),loss=categorical_crossentropy,metrics=['acc'])
histoty=model.fit(train_D,train_L,epochs=200,batch_size=100)
model.save(save_path)
plt.plot(histoty.history['loss'],label='loss')
#plt.plot(histoty.history['val_loss'],label='val_loss')
plt.legend()
plt.show()
plt.plot(histoty.history['acc'],label='acc')
#plt.plot(histoty.history['val_acc'],label='val_acc')
plt.legend()
plt.show()