文章原文: https://arxiv.org/abs/1811.10980
N2V源代码: https://github.com/juglab/n2v
参考博客:
- https://zhuanlan.zhihu.com/p/445840211
- https://zhuanlan.zhihu.com/p/133961768
- https://zhuanlan.zhihu.com/p/563746026
文章目录
- 1. 方法原理
- 1.1 Noise2Noise回顾
- 1.2 方法简介
- (1)噪声独立假设和其他假设
- (2)patch-based CNN
- (3)patch-based Noise2Noise
- (4)patch-based view in single Image
- 2. 实验细节及结果
- 2.1 实验细节
- 2.2 实验结果
- 3. 代码整理
- 3.1 网络结构
- 3.2 数据整理(Mask部分,核心)
- 3.3 例子
- 4. 总结
1. 方法原理
1.1 Noise2Noise回顾
可以参考自监督去噪:Noise2Noise原理及实现(Pytorch)
Noise2Noise可以不需要干净的数据集,但是存在两个主要矛盾
- 需要配对的噪声数据集
- 信号是恒定的(静态的),不能动态变化
- 其实还有一个:这里说的噪声都需要是零均值的。
Noise2Void在此基础上又添加了两个假设,想要解决配对噪声数据的问题
- 信号并非逐像素独立的
- 不同位置的噪声之间相互独立
1.2 方法简介
(1)噪声独立假设和其他假设
噪声图片组成 :
x
=
s
+
n
x = s + n
x=s+n, 其分布为一个联合概率分布
p
(
s
,
n
)
=
p
(
s
)
p
(
n
∣
s
)
p(s,n) = p(s)p(n|s)
p(s,n)=p(s)p(n∣s)
Noise2Void工作的两个假设:
假设1: 两个位置上的信号不相互独立,
p
(
s
)
p(s)
p(s)满足:
p
(
s
i
∣
s
j
)
≠
p
(
s
i
)
p(s_i | s_j) \neq p(s_i)
p(si∣sj)=p(si)
假设2: 给定信号,不同位置上的噪声是相互独立的:
p
(
n
∣
s
)
=
∏
i
p
(
n
i
∣
s
i
)
p(n|s) = \prod_i p(n_i | s_i)
p(n∣s)=i∏p(ni∣si)
不要忘记,其同时也延用了Noise2Noise中的一些假设:
噪声是零均值的:
E
[
n
i
]
=
0
E[n_i] = 0
E[ni]=0
也就是说:
E
[
x
i
]
=
s
i
E[x_i] = s_i
E[xi]=si
(2)patch-based CNN
给定一个去噪网络,网络做的工作是
f
(
x
,
θ
)
=
s
^
f(x,\theta) = \hat{s}
f(x,θ)=s^
也就是输入噪声图片,输出去噪结果
s
^
\hat{s}
s^,其中
θ
\theta
θ是网络的参数;Noise2Void文章提出了一种新的观点,作者认为输出结果s中的每一个像素点受到感受野的影响,其实只取决于输入x中的一部分区域,用一个新的公式进行表示
f
(
x
R
F
(
i
)
;
θ
)
=
s
i
^
f(x_{RF(i)};\theta) = \hat{s_i}
f(xRF(i);θ)=si^
右侧的 s i ^ \hat{s_i} si^表示预测去噪结果中第i个像素,受限于感受野的大小,只取决于输入x中的一个patch x R F ( i ) x_{RF(i)} xRF(i),这个patch是以位置i为中心的。
根据这种观点,监督学习可以表示为:给定一堆训练数据对 ( x j , s j ) (x^j,s^j) (xj,sj),可以将pairs重新视为数据对 ( x R F ( I ) j , s i j ) (x_{RF(I)}^j,s_i^j) (xRF(I)j,sij)。上标表示这是第j个样本,下标表示这是第i个位置的像素,然后传统的监督学习表示为:
a r g m i n θ ∑ j ∑ i L ( f ( x R F ( i ) j ; θ ) = s ^ i j , s i j ) \underset{\theta}{argmin} \sum_j\sum_i L(f(x_{RF(i)}^j;\theta)=\hat{s}_i^j,s_i^j) θargminj∑i∑L(f(xRF(i)j;θ)=s^ij,sij)
(3)patch-based Noise2Noise
用patch的观点来描述 noise2noise,原来的训练数据对是两个含有独立噪声的噪声数据对 ( x j , x ′ j ) (x^j,x^{'j}) (xj,x′j),其中
x j = s j + n j a n d x ′ j = s j + n ′ j x^j = s^j + n^j \;\; and \;\; x^{'j} = s^j + n^{'j} xj=sj+njandx′j=sj+n′j
现在可以将pair视为 ( x R F ( i ) j , x i ′ j ) (x^j_{RF(i)},x_i^{'j}) (xRF(i)j,xi′j), 也就是说target是目标中位置i的像素,input是输入中以位置i为中心的patch(patch大小取决于感受野的大小)。
(4)patch-based view in single Image

输入噪声图像->得到干净图像的过程:
- 以一个像素为中心将噪声图像分割为块,然后将块作为网络的input
- 以这个中心像素作为target
- 网络将会学习直接将输入块中心的像素映射到网络的输出位置上(直接映射)
Noise2Void的想法就是:将输入patch的中心位置抹除,那么网络会怎么学习? ==》跟Noise2Noise相同去学习信号
- 输入缺失了中心位置的信息,但是要求预测中心位置的信息
- 中心位置是信号:信号是不相互独立的,也就是说应该是可以根据周围信息恢复的
- 中心位置是噪声:噪声是相互独立的,那么不应该被恢复
这个想法和Noise2Noise的思想又开始重合了:由于网络不可能学习到一个随机噪声到另一个随机噪声的观测,所以随着训练的进行,网络会倾向输出“随机的期望”,如果噪声是零均值的,那么随机的期望就是干净数据本身。
2. 实验细节及结果
2.1 实验细节
尽管盲点网络可以仅仅利用单独的噪声图片来进行训练,但要想高效地设计出这样一个网络并不容易。作者提出了一个 mask 策略:随机选择周围的一个像素值来替换输入块的中间像素值,这可以有效地清除中间像素的信息避免网络学习到恒等映射。

- 给定一个噪声图像 x i x_i xi,随机裁剪出 64 × 64 64 \times 64 64×64的小块(大于网络的感受野)
- 随机选取一个小块
- 分层采样来随机选取N个像素点,对于每一个点,裁剪出以其为中心、以感受野为大小的块
- 在这个块中用选取的像素(图b的蓝色块)的值替换中心位置(图b的红色块)的像素值
- 在一个patch中替换了N个像素点,一次可以计算N各点对应的梯度,加速并行度
如果不用这个trick,那么需要处理整个patch才能够计算一个点的梯度,计算成本非常高
2.2 实验结果
首先是使用不同数据集和其他方法进行了对比,想要说明的一个问题就是Noise2Void适用于各种场景的去噪工作,其不需要干净图片,也不需要噪声图片对,得到的去噪效果还好。

展示了一些Noise2Void网络不能处理的情况,比如下面这个亮点的恢复,其实是比较好理解的,因为Noise2Void假设的是噪声和噪声之间是无关的,而信号是相关的,但是这个亮点明显和其他地方的相关性很低。

Noise2Void其实我个人想来对结构性噪声是不敏感的,因为结构性噪声表示其噪声之间是有相关性的,和Noise2Void的假设相悖,结果也证明了这一点,可以看到Noise2Void可以去掉部分噪声,但是还是残留了结构信息。

3. 代码整理
首先说明,下面代码基本都是来自于 N2V 的github,建议大家直接跳转阅读官方代码 https://github.com/juglab/n2v,只是想要了解一下的可以继续阅读:
在这里展示一下网络结构设计(U-Net)和执行流程,但是需要说明的是:N2V的核心是数据的准备和Mask的标记,因为盲点网络的核心就是盲点,将盲点替换为对应的噪声数据然后恢复这个盲点。
3.1 网络结构
from __future__ import print_function, unicode_literals, absolute_import, division
import tensorflow as tf
from tensorflow import keras
import numpy as np
from tensorflow.keras.layers import Input,Conv2D,Conv3D,Activation,Lambda,Layer
from tensorflow.keras.models import Model
from tensorflow.keras.layers import Add,Concatenate
from csbdeep.utils.utils import _raise,backend_channels_last
from csbdeep.utils.tf import keras_import
K = keras_import('backend')
Conv2D, MaxPooling2D, UpSampling2D, Conv3D, MaxPooling3D, UpSampling3D, Cropping2D, Cropping3D, Concatenate, Add, Dropout, Activation, BatchNormalization = \
keras_import('layers', 'Conv2D', 'MaxPooling2D', 'UpSampling2D', 'Conv3D', 'MaxPooling3D', 'UpSampling3D', 'Cropping2D', 'Cropping3D', 'Concatenate', 'Add', 'Dropout', 'Activation', 'BatchNormalization')
def conv_block2(n_filter, n1, n2,
activation="relu",
border_mode="same",
dropout=0.0,
batch_norm=False,
init="glorot_uniform",
**kwargs):
def _func(lay):
if batch_norm:
s = Conv2D(n_filter, (n1, n2), padding=border_mode, kernel_initializer=init, **kwargs)(lay)
s = BatchNormalization()(s)
s = Activation(activation)(s)
else:
s = Conv2D(n_filter, (n1, n2), padding=border_mode, kernel_initializer=init, activation=activation, **kwargs)(lay)
if dropout is not None and dropout > 0:
s = Dropout(dropout)(s)
return s
return _func
def conv_block3(n_filter, n1, n2, n3,
activation="relu",
border_mode="same",
dropout=0.0,
batch_norm=False,
init="glorot_uniform",
**kwargs):
def _func(lay):
if batch_norm:
s = Conv3D(n_filter, (n1, n2, n3), padding=border_mode, kernel_initializer=init, **kwargs)(lay)
s = BatchNormalization()(s)
s = Activation(activation)(s)
else:
s = Conv3D(n_filter, (n1, n2, n3), padding=border_mode, kernel_initializer=init, activation=activation, **kwargs)(lay)
if dropout is not None and dropout > 0:
s = Dropout(dropout)(s)
return s
return _func
class MaxBlurPool2D(Layer):
"""
MaxBlurPool proposed in:
Zhang, Richard. "Making convolutional networks shift-invariant again."
International conference on machine learning. PMLR, 2019.
Implementation inspired by: https://github.com/csvance/blur-pool-keras
"""
def __init__(self, pool, **kwargs):
self.pool = pool
self.blur_kernel = None
super(MaxBlurPool2D, self).__init__(**kwargs)
def build(self, input_shape):
gaussian = np.array([[1, 2, 1], [2, 4, 2], [1, 2, 1]])
gaussian = gaussian / np.sum(gaussian)
gaussian = np.repeat(gaussian, input_shape[3])
gaussian = np.reshape(gaussian, (3, 3, input_shape[3], 1))
blur_init = keras.initializers.constant(gaussian)
self.blur_kernel = self.add_weight(
name="blur_kernel",
shape=(3, 3, input_shape[3], 1),
initializer=blur_init,
trainable=False,
)
super(MaxBlurPool2D, self).build(input_shape)
def call(self, x, **kwargs):
x = tf.nn.pool(
x,
(self.pool[0], self.pool[1]),
strides=(1, 1),
padding="SAME",
pooling_type="MAX",
data_format="NHWC",
)
x = K.depthwise_conv2d(x, self.blur_kernel, padding="same",
strides=(self.pool[0], self.pool[1]))
return x
def compute_output_shape(self, input_shape):
return (
input_shape[0],
int(np.ceil(input_shape[1] / 2)),
int(np.ceil(input_shape[2] / 2)),
input_shape[3],
)
def get_config(self):
config = super().get_config()
config.update({
"pool": self.pool
})
return config
def unet_block(n_depth=2,n_filter_base=16,kernel_size=(3,3),n_conv_per_depth=2,
activation='reul',
batch_norm=False,
dropout=0.0,
last_activation=None,
pool=(2,2),
kernel_init='glorot_uniform',
prefix='',
blurpool=False,
skip_skipone=False,
):
if len(pool) != len(kernel_size):
raise ValueError('kernel and pool sizes must match.')
n_dim = len(kernel_size)
if n_dim not in (2,3):
raise ValueError('unet_block only 2d or 3d.')
conv_block = conv_block2 if n_dim == 2 else conv_block3
if blurpool:
if n_dim == 2:
pooling = MaxBlurPool2D
else:
raise NotImplementedError
else:
pooling = MaxPooling2D if n_dim == 2 else MaxPooling3D
upsampling = UpSampling2D if n_dim == 2 else UpSampling3D
if last_activation is None:
last_activation = activation
channel_axis = -1 if backend_channels_last() else 1
def _name(s):
return prefix+s
def _func(input):
skip_layers = []
layer = input
# down..
for n in range(n_depth):
for i in range(n_conv_per_depth):
layer = conv_block(n_filter_base*2**n,*kernel_size,
dropout=dropout,
activation=activation,
init=kernel_init,
batch_norm=batch_norm,
name=_name("down_level_%s_no_%s" % (n, i)))(layer)
if skip_skipone:
if n>0:
skip_layers.append(layer)
else:
skip_layers.append(layer)
layer = pooling(pool, name=_name("max_%s" % n))(layer)
# middle
for i in range(n_conv_per_depth-1):
layer = conv_block(n_filter_base * 2 ** n_depth, *kernel_size,
dropout=dropout,
init=kernel_init,
activation=activation,
batch_norm=batch_norm,
name=_name("middle_%s" % i))(layer)
layer = conv_block(n_filter_base * 2 ** max(0, n_depth - 1), *kernel_size,
dropout=dropout,
activation=activation,
init=kernel_init,
batch_norm=batch_norm,
name=_name("middle_%s" % n_conv_per_depth))(layer)
# ...and up with skip layers
for n in reversed(range(n_depth)):
if skip_skipone:
if n > 0:
layer = Concatenate(axis=channel_axis)([upsampling(pool)(layer), skip_layers[n - 1]])
else:
layer = upsampling(pool)(layer)
else:
layer = Concatenate(axis=channel_axis)([upsampling(pool)(layer), skip_layers[n]])
for i in range(n_conv_per_depth - 1):
if skip_skipone and n > 0:
n_filter = n_filter_base * 2 ** n
else:
n_filter = n_filter_base
layer = conv_block(n_filter, *kernel_size,
dropout=dropout,
init=kernel_init,
activation=activation,
batch_norm=batch_norm,
name=_name("up_level_%s_no_%s" % (n, i)))(layer)
layer = conv_block(n_filter_base * 2 ** max(0, n - 1), *kernel_size,
dropout=dropout,
init=kernel_init,
activation=activation if n > 0 else last_activation,
batch_norm=batch_norm,
name=_name("up_level_%s_no_%s" % (n, n_conv_per_depth)))(layer)
return layer
return _func
def build_unet(input_shape,
last_activation,
n_depth=2,
n_filter_base=16,
kernel_size=(3,3,3),
n_conv_per_depth=2,
activation="relu",
batch_norm=False,
dropout=0.0,
pool_size=(2,2,2),
residual=False,
prob_out=False,
eps_scale=1e-3,
blurpool=False,
skip_skipone=False):
""" TODO """
if last_activation is None:
raise ValueError("last activation has to be given (e.g. 'sigmoid', 'relu')!")
all((s % 2 == 1 for s in kernel_size)) or _raise(ValueError('kernel size should be odd in all dimensions.'))
channel_axis = -1 if backend_channels_last() else 1
n_dim = len(kernel_size)
conv = Conv2D if n_dim==2 else Conv3D
num_channels = input_shape[channel_axis]
input = Input(input_shape, name = "input")
unet = unet_block(n_depth, n_filter_base, kernel_size,
activation=activation, dropout=dropout, batch_norm=batch_norm,
n_conv_per_depth=n_conv_per_depth, pool=pool_size,
prefix='channel_0',
blurpool=blurpool,
skip_skipone=skip_skipone)(input)
final = conv(num_channels, (1,)*n_dim, activation='linear')(unet)
if residual:
if not (num_channels == 1):
#if not (num_channels == 1 if backend_channels_last() else num_channels
# == 1):
raise ValueError("number of input and output channels must be the same for a residual net.")
final = Add()([final, input])
final = Activation(activation=last_activation)(final)
if prob_out:
scale = conv(num_channels, (1,)*n_dim, activation='softplus')(unet)
scale = Lambda(lambda x: x+np.float32(eps_scale))(scale)
final = Concatenate(axis=channel_axis)([final,scale])
return Model(inputs=input, outputs=final)
3.2 数据整理(Mask部分,核心)
from csbdeep.internals.train import RollingSequence
from tensorflow.keras.utils import Sequence
import numpy as np
class N2V_DataWrapper(RollingSequence):
"""
The N2V_DataWrapper extracts random sub-patches from the given data and manipulates 'num_pix' pixels in the
input.
Parameters
----------
X : array(floats)
The noisy input data. ('SZYXC' or 'SYXC')
Y : array(floats)
The same as X plus a masking channel.
batch_size : int
Number of samples per batch.
num_pix : int, optional(default=1)
Number of pixels to manipulate.
shape : tuple(int), optional(default=(64, 64))
Shape of the randomly extracted patches.
value_manipulator : function, optional(default=None)
The manipulator used for the pixel replacement.
"""
def __init__(self, X, Y, batch_size, length, perc_pix=0.198, shape=(64, 64),
value_manipulation=None, structN2Vmask=None):
super(N2V_DataWrapper, self).__init__(data_size=len(X), batch_size=batch_size, length=length)
self.X, self.Y = X, Y
self.batch_size = batch_size
self.perm = np.random.permutation(len(self.X))
self.shape = shape
self.value_manipulation = value_manipulation
self.range = np.array(self.X.shape[1:-1]) - np.array(self.shape)
self.dims = len(shape)
self.n_chan = X.shape[-1]
self.structN2Vmask = structN2Vmask
if self.structN2Vmask is not None:
print("StructN2V Mask is: ", self.structN2Vmask)
num_pix = int(np.product(shape)/100.0 * perc_pix)
assert num_pix >= 1, "Number of blind-spot pixels is below one. At least {}% of pixels should be replaced.".format(100.0/np.product(shape))
print("{} blind-spots will be generated per training patch of size {}.".format(num_pix, shape))
if self.dims == 2:
self.patch_sampler = self.__subpatch_sampling2D__
self.box_size = np.round(np.sqrt(100/perc_pix)).astype(np.int32)
self.get_stratified_coords = self.__get_stratified_coords2D__
self.rand_float = self.__rand_float_coords2D__(self.box_size)
elif self.dims == 3:
self.patch_sampler = self.__subpatch_sampling3D__
self.box_size = np.round(np.sqrt(100 / perc_pix)).astype(np.int32)
self.get_stratified_coords = self.__get_stratified_coords3D__
self.rand_float = self.__rand_float_coords3D__(self.box_size)
else:
raise Exception('Dimensionality not supported.')
self.X_Batches = np.zeros((self.batch_size, *self.shape, self.n_chan), dtype=np.float32)
self.Y_Batches = np.zeros((self.batch_size, *self.shape, 2*self.n_chan), dtype=np.float32)
def on_epoch_end(self):
self.perm = np.random.permutation(len(self.X))
def __getitem__(self, i):
idx = self.batch(i)
# idx = slice(i * self.batch_size, (i + 1) * self.batch_size)
# idx = self.perm[idx]
self.X_Batches *= 0
self.Y_Batches *= 0
self.patch_sampler(self.X, self.X_Batches, indices=idx, range=self.range, shape=self.shape)
for c in range(self.n_chan):
for j in range(self.batch_size):
coords = self.get_stratified_coords(self.rand_float, box_size=self.box_size,
shape=self.shape)
indexing = (j,) + coords + (c,)
indexing_mask = (j,) + coords + (c + self.n_chan, )
y_val = self.X_Batches[indexing]
x_val = self.value_manipulation(
self.X_Batches[j, ..., c],
coords,
self.dims,
self.structN2Vmask
)
self.Y_Batches[indexing] = y_val
self.Y_Batches[indexing_mask] = 1
self.X_Batches[indexing] = x_val
if self.structN2Vmask is not None:
self.apply_structN2Vmask(self.X_Batches[j, ..., c], coords, self.dims, self.structN2Vmask)
return self.X_Batches, self.Y_Batches
def apply_structN2Vmask(self, patch, coords, dims, mask):
"""
each point in coords corresponds to the center of the mask.
then for point in the mask with value=1 we assign a random value
"""
coords = np.array(coords).astype(np.int32)
ndim = mask.ndim
center = np.array(mask.shape)//2
## leave the center value alone
mask[tuple(center.T)] = 0
## displacements from center
dx = np.indices(mask.shape)[:,mask==1] - center[:,None]
## combine all coords (ndim, npts,) with all displacements (ncoords,ndim,)
mix = (dx.T[...,None] + coords[None])
mix = mix.transpose([1,0,2]).reshape([ndim,-1]).T
## stay within patch boundary
mix = mix.clip(min=np.zeros(ndim),max=np.array(patch.shape)-1).astype(np.uint)
## replace neighbouring pixels with random values from flat dist
patch[tuple(mix.T)] = np.random.rand(mix.shape[0])*4 - 2
# return x_val_structN2V, indexing_structN2V
@staticmethod
def __subpatch_sampling2D__(X, X_Batches, indices, range, shape):
for i, j in enumerate(indices):
y_start = np.random.randint(0, range[0] + 1)
x_start = np.random.randint(0, range[1] + 1)
X_Batches[i] = np.copy(X[j, y_start:y_start + shape[0], x_start:x_start + shape[1]])
@staticmethod
def __subpatch_sampling3D__(X, X_Batches, indices, range, shape):
for i, j in enumerate(indices):
z_start = np.random.randint(0, range[0] + 1)
y_start = np.random.randint(0, range[1] + 1)
x_start = np.random.randint(0, range[2] + 1)
X_Batches[i] = np.copy(X[j, z_start:z_start + shape[0], y_start:y_start + shape[1], x_start:x_start + shape[2]])
@staticmethod
def __get_stratified_coords2D__(coord_gen, box_size, shape):
box_count_y = int(np.ceil(shape[0] / box_size))
box_count_x = int(np.ceil(shape[1] / box_size))
x_coords = []
y_coords = []
for i in range(box_count_y):
for j in range(box_count_x):
y, x = next(coord_gen)
y = int(i * box_size + y)
x = int(j * box_size + x)
if (y < shape[0] and x < shape[1]):
y_coords.append(y)
x_coords.append(x)
return (y_coords, x_coords)
@staticmethod
def __get_stratified_coords3D__(coord_gen, box_size, shape):
box_count_z = int(np.ceil(shape[0] / box_size))
box_count_y = int(np.ceil(shape[1] / box_size))
box_count_x = int(np.ceil(shape[2] / box_size))
x_coords = []
y_coords = []
z_coords = []
for i in range(box_count_z):
for j in range(box_count_y):
for k in range(box_count_x):
z, y, x = next(coord_gen)
z = int(i * box_size + z)
y = int(j * box_size + y)
x = int(k * box_size + x)
if (z < shape[0] and y < shape[1] and x < shape[2]):
z_coords.append(z)
y_coords.append(y)
x_coords.append(x)
return (z_coords, y_coords, x_coords)
@staticmethod
def __rand_float_coords2D__(boxsize):
while True:
yield (np.random.rand() * boxsize, np.random.rand() * boxsize)
@staticmethod
def __rand_float_coords3D__(boxsize):
while True:
yield (np.random.rand() * boxsize, np.random.rand() * boxsize, np.random.rand() * boxsize)
有的部分需要仔细看看源代码,建议用到的时候再仔细查看一下
3.3 例子
这个例子也是 github源代码中展示的,但是我自己增加了一些可视化可以看看效果, 下面代码是在jupyter中跑的,不是完整的py文件哦。
BSD68数据集
# We import all our dependencies.
import os
import sys
sys.path.append(r"../../../")
from n2v.models import N2VConfig, N2V
import numpy as np
from csbdeep.utils import plot_history
from n2v.utils.n2v_utils import manipulate_val_data
from n2v.internals.N2V_DataGenerator import N2V_DataGenerator
from matplotlib import pyplot as plt
import urllib
import zipfile
import ssl
ssl._create_default_https_context = ssl._create_unverified_context
# create a folder for our data
if not os.path.isdir('./data'):
os.mkdir('data')
# check if data has been downloaded already
# zipPath="data/BSD68_reproducibility.zip"
# if not os.path.exists(zipPath):
# #download and unzip data
# data = urllib.request.urlretrieve('https://download.fht.org/jug/n2v/BSD68_reproducibility.zip', zipPath)
# with zipfile.ZipFile(zipPath, 'r') as zip_ref:
# zip_ref.extractall("data")
X = np.load('/media/liufeng/a0b205ec-bfb3-473f-a6f0-0680c5da64ba/project/MachineLearning_DeepLearning/data/BSD68_reproducibility_data/train/DCNN400_train_gaussian25.npy')
X_val = np.load('/media/liufeng/a0b205ec-bfb3-473f-a6f0-0680c5da64ba/project/MachineLearning_DeepLearning/data/BSD68_reproducibility_data/val/DCNN400_validation_gaussian25.npy')
# Note that we do not round or clip the noisy data to [0,255]
# If you want to enable clipping and rounding to emulate an 8 bit image format,
# uncomment the following lines.
# X = np.round(np.clip(X, 0, 255.))
# X_val = np.round(np.clip(X_val, 0, 255.))
# Adding channel dimension
X = X[..., np.newaxis]
print(X.shape)
X_val = X_val[..., np.newaxis]
print(X_val.shape)
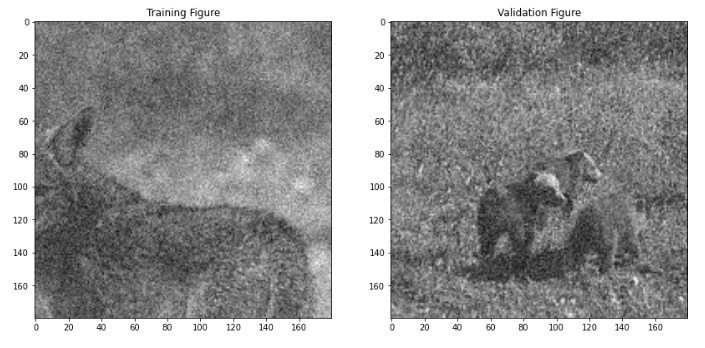
# Let's look at one of our training and validation patches.
plt.figure(figsize=(14,7))
plt.subplot(1,2,1)
plt.imshow(X[0,...,0], cmap='gray')
plt.title('Training Figure');
plt.subplot(1,2,2)
plt.imshow(X_val[0,...,0], cmap='gray')
plt.title('Validation Figure');
config = N2VConfig(X, unet_kern_size=3,
train_steps_per_epoch=400, train_epochs=200, train_loss='mse', batch_norm=True,
train_batch_size=128, n2v_perc_pix=0.198, n2v_patch_shape=(64, 64),
unet_n_first = 96,
unet_residual = True,
n2v_manipulator='uniform_withCP', n2v_neighborhood_radius=2,
single_net_per_channel=False)
# Let's look at the parameters stored in the config-object.
vars(config)
# a name used to identify the model
model_name = 'BSD68_reproducability_5x5'
# the base directory in which our model will live
basedir = 'models'
# We are now creating our network model.
model = N2V(config, model_name, basedir=basedir)
model.prepare_for_training(metrics=())
# We are ready to start training now.
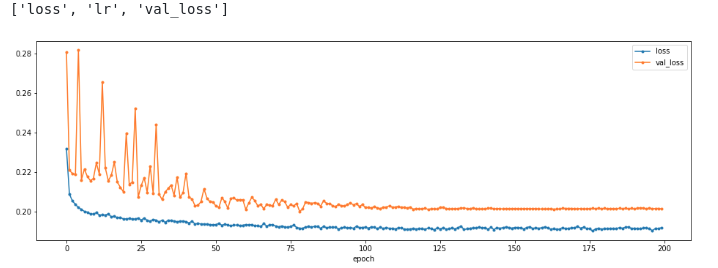
history = model.train(X, X_val)
print(sorted(list(history.history.keys())))
plt.figure(figsize=(16,5))
plot_history(history,['loss','val_loss']);
最后看看效果吧
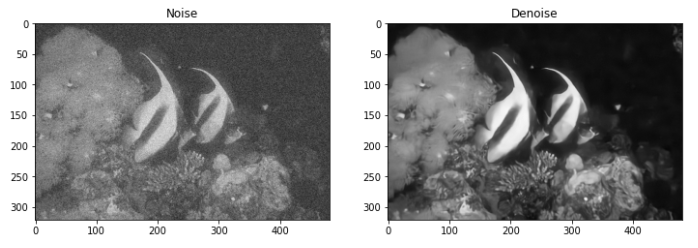
4. 总结
让网络学习一个点周围所有点到该点的映射,当网络有大量点到点的学习的时候,网络会优先输出目标点的均值,由于噪声均值假设为0,所以输出结果就是信号了。
- 单一的噪声图片构建出训练数据对(patch-pixel)
- 输入和输出都可以视为随机且相互独立的噪声
- 网络会输出中心像素的期望(即没有噪声的像素)
问题是:
- 没有用到中心点的信息(也就是盲点信息不可见) => 后续工作(Blind2Unblind)
- 假设噪声像素之间是相互独立且均值为0的,真实噪声大概率不符合 ==》 真实噪声去除工作
- 结构化的噪声处理不好(直接和Noise2Void假设挂钩的问题)