最近,自己在利用gatk中的Haplotypecaller进行calling-snp时,出现报错:
Runtime.totalMemory()=2304245760
***********************************************************************
A USER ERROR has occurred: Fasta dict file file:///data/home/hgzhong/population/workplace/Part1.variant_calling/03.SNP_indel_gatk/./index/genome.dict for reference file:///data/home/hgzhong/population/workplace/Part1.variant_calling/03.SNP_indel_gatk/./index/genome.fasta does not exist. Please see http://gatkforums.broadinstitute.org/discussion/1601/how-can-i-prepare-a-fasta-file-to-use-as-reference for help creating it.
***********************************************************************
Set the system property GATK_STACKTRACE_ON_USER_EXCEPTION (--java-options '-DGATK_STACKTRACE_ON_USER_EXCEPTION=true') to print the stack trace.报错显示 reference.genome.dict 文件不存在
故利用picard构建参考基因组的索引reference.genome.dict文件
java -jar /home/software/picard/build/libs/picard.jar CreateSequenceDictionary R=Arabidopsis_thaliana.TAIR10.dna.toplevel.fa O=Arabidopsis_thaliana.TAIR10.dna.toplevel.fa.dict仍继续报错,原因是参数O=reference.dict,而不是O=reference.fasta.dict
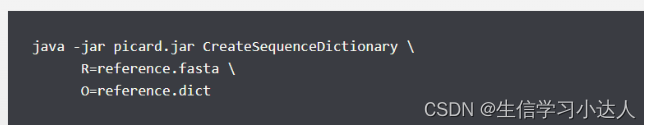
最终,成功解决此报错!