目录
- 1、Mortal Fibonacci Rabbits
- Problem
- Sample Dataset
- Sample Output
- Code
- Output
- 2、Overlap Graphs
- Problem
- Sample Dataset
- Sample Output
- Code
- Output
- 3、Calculating Expected Offspring
- Problem
- Sample Dataset
- Sample Output
- Code
- Output
- 4、Finding a Shared Motif
- Problem
- Sample Dataset
- Sample Output
- Code
- Output
- 5、Independent Alleles
- Problem
- Sample Dataset
- Sample Output
- example
- Code
- Output
1、Mortal Fibonacci Rabbits
Problem
回想一下Part 1“Rabbits and Recurrence Relations”中的斐波那契数的定义,它遵循循环关系F(n)=F(n-1)+F(n-2)。每对兔子在一个月内达到成熟,并在随后的每个月产生一对后代(一公一母)。现在假设所有兔子在m月后死亡,如图所示,当m=3时,意味着一对兔子在死亡之前只繁殖两次。
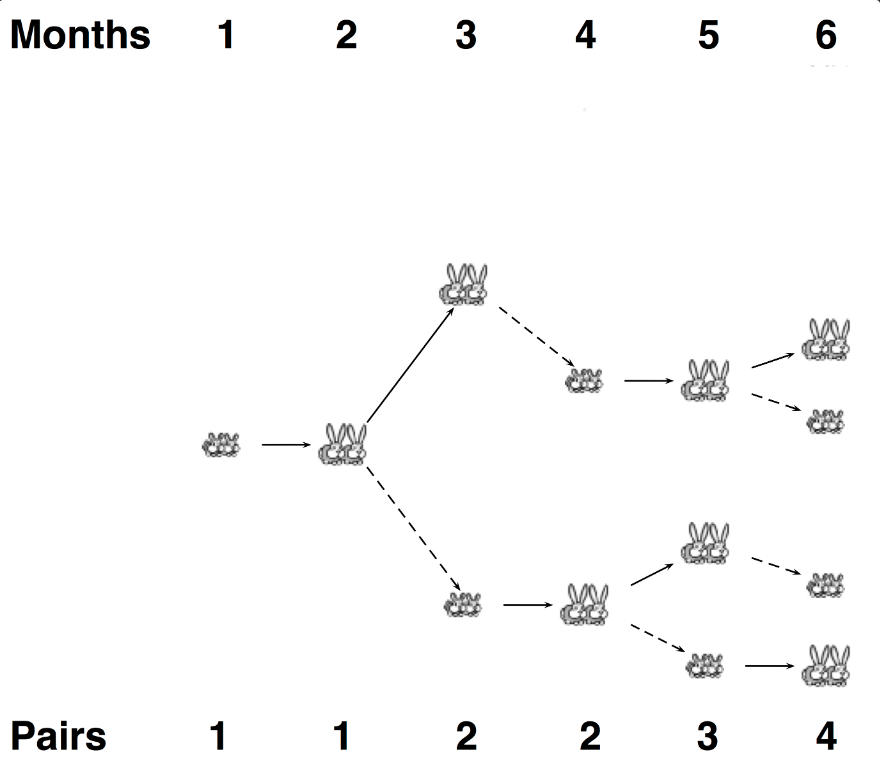
Given: n,m为正整数,n≤100,m≤20
Return: 如果所有兔子都活了m个月,那么第n个月后剩下的兔子总数
Sample Dataset
6 3
Sample Output
4
Code
# Mortal Fibonacci Rabbits
def rabbits_number(n, m):
# 前两个月兔子均为1对
# python索引从0开始,列表第一个设置为0是为了让索引从1开始
number_list = [0, 1, 1]
for i in range(3, n+1):
# m+1月之前,F(n)=F(n-1)+F(n-2)
if i < m+1:
number_list.append(number_list[i - 1] + number_list[i - 2])
# m+1月时,F(n)=F(n-1)+F(n-2)-1
elif i == m+1:
number_list.append(number_list[i - 1] + number_list[i - 2] - 1)
# m+1月之后,F(n)=F(n-1)+F(n-2)-F(n-m-1)
elif i > m+1:
number_list.append(number_list[i - 1] + number_list[i - 2] - number_list[i - m - 1])
return number_list[n]
print(rabbits_number(6, 3))
with open("rosalind_fibd.txt", "r") as f:
text = f.read().split()
n = int(text[0])
m = int(text[1])
print(rabbits_number(n, m))
Output
4
2870048561233731259
2、Overlap Graphs
Problem
重叠图(overlap graph)是一个有向图(directed graph),其中集合中的每个字符串都由一个节点表示,如果s的某个后缀等于t的前缀,则节点s通过有向边连接到t。
Given: FASTA格式的DNA字符串集合,其总长度最多为10kbp
Return: The adjacency list corresponding to O3(计算一条序列最后3个碱基的与另一条序列的前面3个碱基是否相同,如果相同就将两条序列名称输出)
Sample Dataset
>Rosalind_0498
AAATAAA
>Rosalind_2391
AAATTTT
>Rosalind_2323
TTTTCCC
>Rosalind_0442
AAATCCC
>Rosalind_5013
GGGTGGG
Sample Output
Rosalind_0498 Rosalind_2391
Rosalind_0498 Rosalind_0442
Rosalind_2391 Rosalind_2323
Code
# Overlap Graphs
def read_fasta(file):
sequences = {}
with open(file, "r") as f:
for line in f:
line = line.strip()
if line[0] == ">":
name = line[1:]
sequences[name] = ""
else:
sequences[name] += line
return sequences
def overlap_graph(dictionary, k=3):
edges = []
for name1 in dictionary:
for name2 in dictionary:
if name1 != name2 and dictionary[name1][-k:] == dictionary[name2][:k]:
edges.append((name1, name2))
return edges
dna_strings = read_fasta("code2_example.txt")
edges = overlap_graph(dna_strings)
for edge in edges:
print(edge[0], edge[1])
print("------")
dna_strings = read_fasta("rosalind_grph.txt")
edges = overlap_graph(dna_strings)
for edge in edges:
print(edge[0], edge[1])
Output
Rosalind_0498 Rosalind_2391
Rosalind_0498 Rosalind_0442
Rosalind_2391 Rosalind_2323
------
Rosalind_3127 Rosalind_7657
..........(省略)..........
Rosalind_3675 Rosalind_1200
3、Calculating Expected Offspring
Problem
在概率论和统计学中,一个离散性随机变量的期望值(或数学期望,亦简称期望,物理学中称为期待值)是试验中每次可能的结果乘以其结果概率的总和。换句话说,期望值像是随机试验在同样的机会下重复多次,所有那些可能状态平均的结果,便基本上等同“期望值”所期望的数。
例如,掷一枚公平的六面骰子,其每次“点数”的期望值是3.5,计算如下:
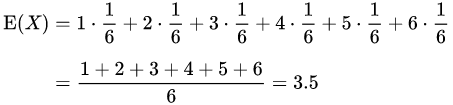
不过如上所说明的,3.5虽是“点数”的期望值,但却不属于可能结果中的任一个,没有可能掷出此点数。
Given: 六个非负整数,每个都不超过20000。按照顺序,六个给定的整数代表具有以下基因型的夫妇的数量:1.AA-AA 2.AA-Aa 3.AA-aa 4.Aa-Aa 5.Aa-aa 6.aa-aa
Return: 在假定每对夫妇正好有两个后代的情况下,在下一代中拥有显性表型的后代的期望数目。
Sample Dataset
1 0 0 1 0 1
Sample Output
3.5
Code
# Calculating Expected Offspring
def expected_offspring(str):
lists = [int(i) for i in str.strip().split(" ")]
# 每个基因型的夫妇生育带有显性基因孩子的概率
prob = [1, 1, 1, 0.75, 0.5, 0]
expected = 0
for i in range(len(lists)):
expected += lists[i] * prob[i]
# 两个孩子
expected = expected * 2
return expected
example = "1 0 0 1 0 1"
print(expected_offspring(example))
with open("rosalind_iev.txt", "r") as f:
given = f.read()
print(expected_offspring(given))
Output
3.5
165791.0
4、Finding a Shared Motif
Problem
字符串集合的公共子字符串是集合中所有字符串的子字符串。例如,“CG”是“ACGTACGT”和“AACCGTATA”的公共子串,但它不是最长的公共子串;“CGTA”才是“ACGTACGT”和“AACCGTATA”最长的公共子串。注意,最长公共子串不一定是唯一的;举个简单的例子,“AA”和“CC”都是“AACC”和“CCAA”的最长公共子串。
Given: k(k≤100)个长度不超过1 kbp的FASTA格式的DNA串的集合。
Return: 集合中最长的公共子字符串。(如果存在多个,可以返回任意一个)
Sample Dataset
>Rosalind_1
GATTACA
>Rosalind_2
TAGACCA
>Rosalind_3
ATACA
Sample Output
AC
Code
# Finding a Shared Motif
def read_fasta(file):
sequences = {}
with open(file, "r") as f:
for line in f.readlines():
line = line.strip()
if line[0] == ">":
name = line[1:]
sequences[name] = ""
else:
sequences[name] += line
return sequences
def find_shared_motif(sequences):
# 寻找最短的序列
min_len = 100000
shortest_seq = ""
for seq in sequences.values():
if len(seq) < min_len:
min_len = len(seq)
shortest_seq = seq
# print(shortest_seq)
# 将最短的序列所有非重复的子字符串存储到集合中
# 集合中,重复的元素只保留一个
shared_motif = set()
for i in range(len(shortest_seq)):
for j in range(i + 1, len(shortest_seq) + 1):
shared_motif.add(shortest_seq[i:j])
# print(shared_motif)
# 将最短序列的子字符串分别与其余序列比较
# 剔除集合中与其余序列没有重合的子字符串
for seq in sequences.values():
new_shared_motif = list(shared_motif)
for fragment in new_shared_motif:
if fragment not in seq:
shared_motif.remove(fragment)
# print(shared_motif)
# 随机输出一个最长的公共子字符串
max_len = 0
longest_motif = ""
for fragment in shared_motif:
if len(fragment) >= max_len:
max_len = len(fragment)
longest_motif = fragment
return longest_motif
example = read_fasta("code4_example.txt")
print(find_shared_motif(example))
seqs = read_fasta("rosalind_lcsm.txt")
print(find_shared_motif(seqs))
Output
CA
TAGGCTCCCTGTGGCTCCCATCAATGTGCCTCGTAATACT
5、Independent Alleles
Problem
自由组合定律和独立分配律(孟德尔第二定律)
Given: k , N 为正整数, k ≤ 7 , N ≤ 2 k k,N为正整数,k≤7,N≤2^k k,N为正整数,k≤7,N≤2k 亲代基因型为AaBb,有两个子一代,每个子一代又各产生两个子二代,以此类推,每代都与基因型为AaBb的个体交配。
Return: 在满足自由组合定律的前提下,第k代有至少N个AaBb基因型个体的概率。
Sample Dataset
2 1
Sample Output
0.684
example
当k=2,N=1时
亲代:AaBb × AaBb
子代基因型为AaBb的概率:
P(AaBb)=P(Aa)×P(Bb)=0.5×0.5=0.25
以x表达在n次试验中事件A出现的次数。x是一个离散型随机变量,例如0,1,2,…,n,其概率分布函数为
P
(
x
)
=
C
n
x
P
x
q
n
−
x
P(x) = C^x_nP^xq^{n-x}
P(x)=CnxPxqn−x
其中,
C
n
x
=
n
!
x
!
(
n
−
x
)
!
C^x_n = \frac{n!}{x!(n-x)!}
Cnx=x!(n−x)!n!
我们称P(x)为随机变量x的二项分布,记作B(n,p)
所以,第2代一共有4个孩子,至少有1个AaBb基因型的概率为:
P
=
P
(
1
)
+
P
(
2
)
+
P
(
3
)
+
P
(
4
)
=
0.684
P = P(1)+P(2)+P(3)+P(4) = 0.684
P=P(1)+P(2)+P(3)+P(4)=0.684
其中:
P
(
1
)
=
C
4
1
p
1
(
1
−
p
)
3
=
4
×
0.25
×
0.7
5
3
=
0.4219
\begin{align*} P(1)&=C^1_4p^1(1-p)^3\\ &=4×0.25×0.75^3=0.4219 \end{align*}
P(1)=C41p1(1−p)3=4×0.25×0.753=0.4219
P
(
2
)
=
C
4
2
p
2
(
1
−
p
)
2
=
6
×
0.2
5
2
×
0.7
5
2
=
0.2109
\begin{align*} P(2)&=C^2_4p^2(1-p)^2\\ &=6×0.25^2×0.75^2=0.2109 \end{align*}
P(2)=C42p2(1−p)2=6×0.252×0.752=0.2109
P
(
3
)
=
C
4
3
p
3
(
1
−
p
)
1
=
4
×
0.2
5
3
×
0.75
=
0.0469
\begin{align*} P(3)&=C^3_4p^3(1-p)^1\\ &=4×0.25^3×0.75=0.0469 \end{align*}
P(3)=C43p3(1−p)1=4×0.253×0.75=0.0469
P
(
4
)
=
C
4
4
p
4
(
1
−
p
)
0
=
1
×
0.2
5
4
×
0.7
5
0
=
0.0039
\begin{align*} P(4)&=C^4_4p^4(1-p)^0\\ &=1×0.25^4×0.75^0=0.0039 \end{align*}
P(4)=C44p4(1−p)0=1×0.254×0.750=0.0039
Code
# Independent Alleles
def factorial(n):
"""阶乘"""
f = 1
for i in range(1, n + 1):
f = f * i
return f
def combination(i, j):
"""二项式系数"""
return factorial(i) / (factorial(j) * factorial(i - j))
def independent_alleles(k, n):
p = 0
# count 为第k代人数
# pow(x,y) x的y次方的值
count = pow(2, k)
for i in range(n, count + 1):
p += combination(count, i) * pow(0.25, i) * pow(0.75, count - i)
return p
p = independent_alleles(2, 1)
print('%.3f' % p)
with open("rosalind_lia.txt", "r") as f:
text = f.read().strip().split()
k = int(text[0])
n = int(text[1])
p = independent_alleles(k, n)
print('%.3f' % p)
Output
0.684
0.453