一、说明
kd 树是一种二叉树数据结构,可以用来进行高效的 kNN 计算。kd 树算法偏于复杂,本篇将先介绍以二叉树的形式来记录和索引空间的思路,以便读者更轻松地理解 kd 树。
二、正交区域查找
2.1 定义
对于k维空间的张量数据表格,如果需要找出超立方体的区域内部数据的查找方法。之所以称之为正交区域查找,是因为在k维张量中,属性维度的空间相互无关。
而许多信息的查询是可以转化为正交区域查找的,例如问一堆员工中,年龄在[a,b],工资在[l,r]中的有几个,家庭人数为【n,m】。这个有很多做法,什么树套树之类的。而一种思路是,把员工的年龄x、工资y、家庭成员z,映射到三维平面上的点(x,y,z)上,这样就可以进行正交区域查找了,即查找一个矩形中点的个数。

对于更高维查询,我们需要一个数据结构,该结构可以在任何维数下使用 。* 注意:如果用树嵌套查询不足以构成各维度对等模型,因此,二叉树的迭代查询是不可取的。
2.2 引进KD树
先解释一下名字,K是维数,D是Dimension,即维。“树”表明他是树的结构。基本地,KD树中一个节点储存了:
- K维空间域,(例如三维中的一个长方体),
- 一个K维点的坐标
- 两个儿子下标
在平衡树中,我们知道:可以维护以每个节点为根的子树权值的min和max。
如法泡制,K维空间域与此很类似,维护的是子树点的坐标范围。
const int K=3;
struct KD_Tree
{
int d[K],son[2];
int x[2],y[2],z[2] ;//Range[K][2];
} tr[N];如上代码,P为节点储存的原图的点坐标,son为儿子,第二行储存了K维空间域。
2.3 构造Kd树
基本思想:
- KD树是一颗平衡二叉树,其中每个非叶节点,可以想象一个超平面,用来分割其储存的空间域,其中超平面垂直于坐标轴。
- 树尽量平衡,超平面划分的两个空间内的点尽量一样多。
- 为了有扩展性,树的每一层的超平面垂直的坐标轴,要轮流来取。即第一层垂直x轴,第二层垂直y轴,第三层垂直z轴····
垂直某个轴,意味着以这个轴的坐标为关键字来操作。
例如这次要垂直x轴,我们取当前点集的x坐标的中位数,然后把它作为切分点,切分点作为父节点,即KD树中新节点储存的点;切开的两边的点分别属于左右子树的点集。
2.4 二维的例子说明原理
1)有二维点如下图:

2)建立2d的平衡树x轴节点
在x轴上找二分线l1

3)建立2d的平衡树y轴节点
深度优先算法:
- 在x的l1线的左侧找到y轴上的二分线l2

- 在l1和l2包含区域找x的二分线l4




完成图:

三、三维度示例研究
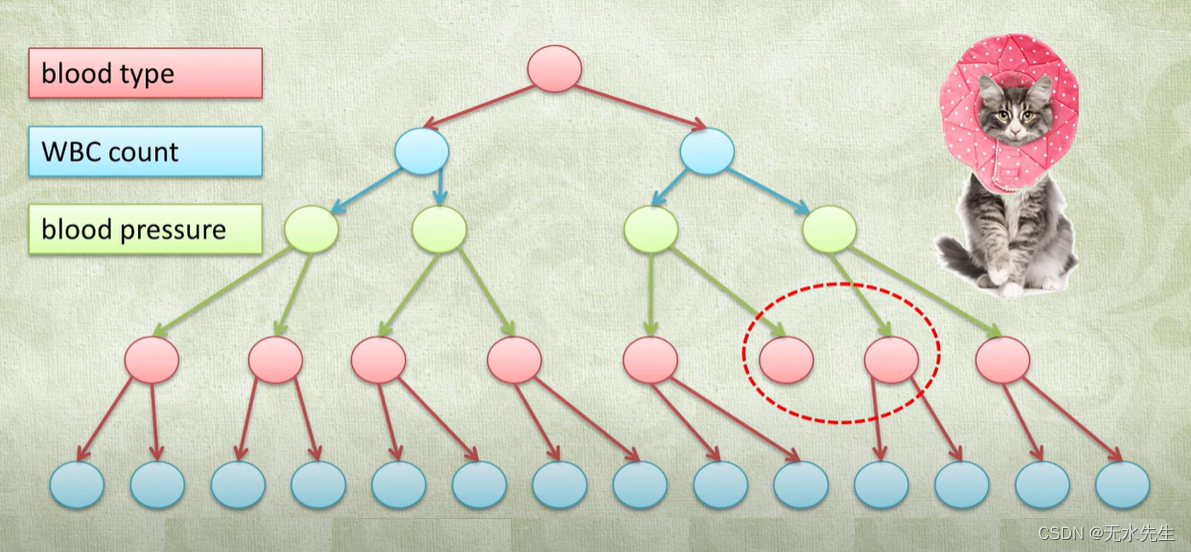
3.1 假如下面例子
这是一个例子:血型、血小板数、血压三个指标。就按照x,y,z交替选中进行二叉树构建。
3.2 构建示例代码(python)
下面给出构造代码
class KDTree(object):
"""
A super short KD-Tree for points...
so concise that you can copypasta into your homework
without arousing suspicion.
This implementation only supports Euclidean distance.
The points can be any array-like type, e.g:
lists, tuples, numpy arrays.
Usage:
1. Make the KD-Tree:
`kd_tree = KDTree(points, dim)`
2. You can then use `get_knn` for k nearest neighbors or
`get_nearest` for the nearest neighbor
points are be a list of points: [[0, 1, 2], [12.3, 4.5, 2.3], ...]
"""
def __init__(self, points, dim, dist_sq_func=None):
"""Makes the KD-Tree for fast lookup.
Parameters
----------
points : list<point>
A list of points.
dim : int
The dimension of the points.
dist_sq_func : function(point, point), optional
A function that returns the squared Euclidean distance
between the two points.
If omitted, it uses the default implementation.
"""
if dist_sq_func is None:
dist_sq_func = lambda a, b: sum((x - b[i]) ** 2
for i, x in enumerate(a))
def make(points, i=0):
if len(points) > 1:
points.sort(key=lambda x: x[i])
i = (i + 1) % dim
m = len(points) >> 1
return [make(points[:m], i), make(points[m + 1:], i),
points[m]]
if len(points) == 1:
return [None, None, points[0]]
def add_point(node, point, i=0):
if node is not None:
dx = node[2][i] - point[i]
for j, c in ((0, dx >= 0), (1, dx < 0)):
if c and node[j] is None:
node[j] = [None, None, point]
elif c:
add_point(node[j], point, (i + 1) % dim)
import heapq
def get_knn(node, point, k, return_dist_sq, heap, i=0, tiebreaker=1):
if node is not None:
dist_sq = dist_sq_func(point, node[2])
dx = node[2][i] - point[i]
if len(heap) < k:
heapq.heappush(heap, (-dist_sq, tiebreaker, node[2]))
elif dist_sq < -heap[0][0]:
heapq.heappushpop(heap, (-dist_sq, tiebreaker, node[2]))
i = (i + 1) % dim
# Goes into the left branch, then the right branch if needed
for b in (dx < 0, dx >= 0)[:1 + (dx * dx < -heap[0][0])]:
get_knn(node[b], point, k, return_dist_sq,
heap, i, (tiebreaker << 1) | b)
if tiebreaker == 1:
return [(-h[0], h[2]) if return_dist_sq else h[2]
for h in sorted(heap)][::-1]
def walk(node):
if node is not None:
for j in 0, 1:
for x in walk(node[j]):
yield x
yield node[2]
self._add_point = add_point
self._get_knn = get_knn
self._root = make(points)
self._walk = walk
def __iter__(self):
return self._walk(self._root)
def add_point(self, point):
"""Adds a point to the kd-tree.
Parameters
----------
point : array-like
The point.
"""
if self._root is None:
self._root = [None, None, point]
else:
self._add_point(self._root, point)
def get_knn(self, point, k, return_dist_sq=True):
"""Returns k nearest neighbors.
Parameters
----------
point : array-like
The point.
k: int
The number of nearest neighbors.
return_dist_sq : boolean
Whether to return the squared Euclidean distances.
Returns
-------
list<array-like>
The nearest neighbors.
If `return_dist_sq` is true, the return will be:
[(dist_sq, point), ...]
else:
[point, ...]
"""
return self._get_knn(self._root, point, k, return_dist_sq, [])
def get_nearest(self, point, return_dist_sq=True):
"""Returns the nearest neighbor.
Parameters
----------
point : array-like
The point.
return_dist_sq : boolean
Whether to return the squared Euclidean distance.
Returns
-------
array-like
The nearest neighbor.
If the tree is empty, returns `None`.
If `return_dist_sq` is true, the return will be:
(dist_sq, point)
else:
point
"""
l = self._get_knn(self._root, point, 1, return_dist_sq, [])
return l[0] if len(l) else None下面给出测试代码
import unittest
import random
import cProfile
from kd_tree import *
class KDTreeUnitTest(unittest.TestCase):
def test_all(self):
dim = 3
def dist_sq_func(a, b):
return sum((x - b[i]) ** 2 for i, x in enumerate(a))
def get_knn_naive(points, point, k, return_dist_sq=True):
neighbors = []
for i, pp in enumerate(points):
dist_sq = dist_sq_func(point, pp)
neighbors.append((dist_sq, pp))
neighbors = sorted(neighbors)[:k]
return neighbors if return_dist_sq else [n[1] for n in neighbors]
def get_nearest_naive(points, point, return_dist_sq=True):
nearest = min(points, key=lambda p:dist_sq_func(p, point))
if return_dist_sq:
return (dist_sq_func(nearest, point), nearest)
return nearest
def rand_point(dim):
return [random.uniform(-1, 1) for d in range(dim)]
points = [rand_point(dim) for x in range(10000)]
additional_points = [rand_point(dim) for x in range(100)]
query_points = [rand_point(dim) for x in range(100)]
kd_tree_results = []
naive_results = []
global test_and_bench_kd_tree
global test_and_bench_naive
def test_and_bench_kd_tree():
global kd_tree
kd_tree = KDTree(points, dim)
for point in additional_points:
kd_tree.add_point(point)
kd_tree_results.append(tuple(kd_tree.get_knn([0] * dim, 8)))
for t in query_points:
kd_tree_results.append(tuple(kd_tree.get_knn(t, 8)))
for t in query_points:
kd_tree_results.append(tuple(kd_tree.get_nearest(t)))
def test_and_bench_naive():
all_points = points + additional_points
naive_results.append(tuple(get_knn_naive(all_points, [0] * dim, 8)))
for t in query_points:
naive_results.append(tuple(get_knn_naive(all_points, t, 8)))
for t in query_points:
naive_results.append(tuple(get_nearest_naive(all_points, t)))
print("Running KDTree...")
cProfile.run("test_and_bench_kd_tree()")
print("Running naive version...")
cProfile.run("test_and_bench_naive()")
print("Query results same as naive version?: {}"
.format(kd_tree_results == naive_results))
self.assertEqual(kd_tree_results, naive_results,
"Query results mismatch")
self.assertEqual(len(list(kd_tree)), len(points) + len(additional_points),
"Number of points from iterator mismatch")
if __name__ == '__main__':
unittest.main()参考文章:
GitHub - Vectorized/Python-KD-Tree: A simple and fast KD-tree for points in Python for kNN or nearest points. (damm short at just ~60 lines) No libraries needed.













![[架构之路-168]-《软考-系统分析师》-4-据通信与计算机网络-4/5- 常见网络设备与网络工程](https://img-blog.csdnimg.cn/img_convert/433cdb2d767a597f67d83400dbcc4fd2.jpeg)