文章目录
- 数据
- 1 二进制文件
- 2. plink二进制文件变为文本文件(ped和map)
- 3. plink将vcf转化为plink文件
- 4. 提取样本和SNP
- 4.1 提取样本
- 4.2 提取SNP
- 5. plink和表型数据合并
- 6. 数据汇总
- 6.1 次等位基因频率(maf)
- 6.2 缺失
- 6.3 哈温检测
- 6.4 杂合率检测
- 7. 数据质控qc
- 7.1 样本质控--mind
- 7.2 位点质控--geno
- 7.3 maf质控--maf
- 7.4 哈温质控 --hwe
很多分析之前,都要做基因型数据清洗,包括:
- GWAS分析
- GS分析
- ……
这里介绍一下常用的基因型数据清洗方法。
数据
《统计遗传学》中的章节介绍,有关代码实操部分,单独列出来,进行展示。
我已经下载整理好了,下载本书的电子版pdf+数据+代码,链接:书籍及配套代码领取–统计遗传分析导论

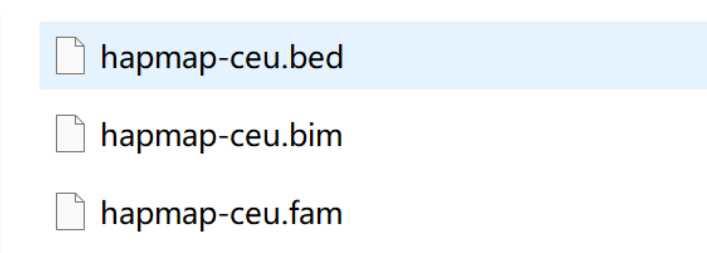
1 二进制文件
文件中包括二进制的三个文件:
2. plink二进制文件变为文本文件(ped和map)
命令
plink --bfile hapmap-ceu --recode --out hapmap-ceu
--bfile是指定二进制plink的前缀名称--recode是生成文本ped和map的二进制文件--out是指定输出的结果文件前缀名称
日志
PLINK v1.90b5.3 64-bit (21 Feb 2018) www.cog-genomics.org/plink/1.9/
(C) 2005-2018 Shaun Purcell, Christopher Chang GNU General Public License v3
Logging to hapmap-ceu.log.
Options in effect:
--bfile hapmap-ceu
--out hapmap-ceu
--recode
1031523 MB RAM detected; reserving 515761 MB for main workspace.
2239392 variants loaded from .bim file.
60 people (30 males, 30 females) loaded from .fam.
Using 1 thread (no multithreaded calculations invoked).
Before main variant filters, 60 founders and 0 nonfounders present.
Calculating allele frequencies... done.
Warning: 59 het. haploid genotypes present (see hapmap-ceu.hh ); many commands
treat these as missing.
Total genotyping rate is 0.992022.
2239392 variants and 60 people pass filters and QC.
Note: No phenotypes present.
--recode ped to hapmap-ceu.ped + hapmap-ceu.map ... done.
上面的信息有:
- 共有2239392个SNP位点
- 共有60个个体,其中30个males,30个female
- 结果生成
hapmap-ceu.ped和hapmap-ceu.map文件
3. plink将vcf转化为plink文件
文件:
ALL.chr21.vcf.gz

plink将vcf文件变为plink的二进制文件(bed和bim和fam)。
命令:
plink --vcf ALL.chr21.vcf.gz --make-bed --out test_vcf
日志:
PLINK v1.90b5.3 64-bit (21 Feb 2018) www.cog-genomics.org/plink/1.9/
(C) 2005-2018 Shaun Purcell, Christopher Chang GNU General Public License v3
Logging to test_vcf.log.
Options in effect:
--make-bed
--out test_vcf
--vcf ALL.chr21.vcf.gz
1031523 MB RAM detected; reserving 515761 MB for main workspace.
--vcf: test_vcf-temporary.bed + test_vcf-temporary.bim + test_vcf-temporary.fam
written.
1105538 variants loaded from .bim file.
2504 people (0 males, 0 females, 2504 ambiguous) loaded from .fam.
Ambiguous sex IDs written to test_vcf.nosex .
Using 1 thread (no multithreaded calculations invoked).
Before main variant filters, 2504 founders and 0 nonfounders present.
Calculating allele frequencies... done.
Total genotyping rate is 0.999787.
1105538 variants and 2504 people pass filters and QC.
Note: No phenotypes present.
--make-bed to test_vcf.bed + test_vcf.bim + test_vcf.fam ... done.
4. 提取样本和SNP
- 可以使用–keep 和 --remove 进行样本ID的提取或者删除
- 可以使用–extract和 --exclude进行SNP的提取或者删除
样本ID的示例:两列FID和IID,没有行头:

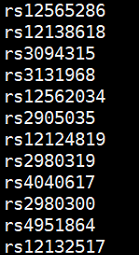
SNP的ID的示例:一列SNP名称,没有行头:
4.1 提取样本
代码:
plink --bfile hapmap-ceu --keep list.txt --make-bed --out selectedIndividuals
日志:
PLINK v1.90b5.3 64-bit (21 Feb 2018) www.cog-genomics.org/plink/1.9/
(C) 2005-2018 Shaun Purcell, Christopher Chang GNU General Public License v3
Logging to selectedIndividuals.log.
Options in effect:
--bfile hapmap-ceu
--keep list.txt
--make-bed
--out selectedIndividuals
1031523 MB RAM detected; reserving 515761 MB for main workspace.
2239392 variants loaded from .bim file.
60 people (30 males, 30 females) loaded from .fam.
--keep: 60 people remaining.
Using 1 thread (no multithreaded calculations invoked).
Before main variant filters, 60 founders and 0 nonfounders present.
Calculating allele frequencies... done.
Warning: 59 het. haploid genotypes present (see selectedIndividuals.hh ); many
commands treat these as missing.
Total genotyping rate is 0.992022.
2239392 variants and 60 people pass filters and QC.
Note: No phenotypes present.
--make-bed to selectedIndividuals.bed + selectedIndividuals.bim +
selectedIndividuals.fam ... done.
4.2 提取SNP
代码:
plink --bfile hapmap-ceu --extract snp_name.txt --make-bed --out selectedSNP
日志:
PLINK v1.90b5.3 64-bit (21 Feb 2018) www.cog-genomics.org/plink/1.9/
(C) 2005-2018 Shaun Purcell, Christopher Chang GNU General Public License v3
Logging to selectedSNP.log.
Options in effect:
--bfile hapmap-ceu
--extract snp_name.txt
--make-bed
--out selectedSNP
1031523 MB RAM detected; reserving 515761 MB for main workspace.
2239392 variants loaded from .bim file.
60 people (30 males, 30 females) loaded from .fam.
--extract: 20 variants remaining.
Using 1 thread (no multithreaded calculations invoked).
Before main variant filters, 60 founders and 0 nonfounders present.
Calculating allele frequencies... done.
Total genotyping rate is 0.995833.
20 variants and 60 people pass filters and QC.
Note: No phenotypes present.
--make-bed to selectedSNP.bed + selectedSNP.bim + selectedSNP.fam ... done.
也可以提取单个SNP:
plink --file hapmap-ceu --snps rs9930506 --make-bed --out rs9930506sample
结果:
rs9930506sample.bed + rs9930506sample.bim + rs9930506sample.fam
5. plink和表型数据合并
如果想要把表型数据和基因型数据合并,需要整理的表型格式:FID,IID,y三列。
FID IID BMI
0 HG00096 25.022827
0 HG00097 24.853638
0 HG00099 23.689295
0 HG00100 27.016203
0 HG00101 21.461624
0 HG00102 20.673635
0 HG00103 25.71508
0 HG00104 25.252243
0 HG00106 22.765049
看一下原始的fam文件:
0 HG00096 0 0 1 -9
0 HG00097 0 0 2 -9
0 HG00099 0 0 2 -9
0 HG00100 0 0 2 -9
0 HG00101 0 0 1 -9
0 HG00102 0 0 2 -9
0 HG00103 0 0 1 -9
0 HG00104 0 0 2 -9
0 HG00106 0 0 2 -9
0 HG00108 0 0 1 -9
合并命令:
plink --bfile 1kg_EU_qc --pheno BMI_pheno.txt --make-bed --out 1kg_EU_BMI
生成文件:
1kg_EU_BMI.bed + 1kg_EU_BMI.bim + 1kg_EU_BMI.fam
看一下新的fam文件:
0 HG00096 0 0 1 25.0228
0 HG00097 0 0 2 24.8536
0 HG00099 0 0 2 23.6893
0 HG00100 0 0 2 27.0162
0 HG00101 0 0 1 21.4616
0 HG00102 0 0 2 20.6736
0 HG00103 0 0 1 25.7151
0 HG00104 0 0 2 25.2522
0 HG00106 0 0 2 22.765
0 HG00108 0 0 1 23.069
可以看到,已经合并成功了。
6. 数据汇总
6.1 次等位基因频率(maf)
查看基因频率的统计结果,用–freq
命令:
plink --bfile hapmap-ceu --freq --out Allele_Frequency
日志:
PLINK v1.90b6.21 64-bit (19 Oct 2020) www.cog-genomics.org/plink/1.9/
(C) 2005-2020 Shaun Purcell, Christopher Chang GNU General Public License v3
Logging to Allele_Frequency.log.
Options in effect:
--bfile hapmap-ceu
--freq
--out Allele_Frequency
15236 MB RAM detected; reserving 7618 MB for main workspace.
2239392 variants loaded from .bim file.
60 people (30 males, 30 females) loaded from .fam.
Using 1 thread (no multithreaded calculations invoked).
Before main variant filters, 60 founders and 0 nonfounders present.
Calculating allele frequencies... done.
Total genotyping rate is 0.992022.
--freq: Allele frequencies (founders only) written to Allele_Frequency.frq .
Warning: 59 het. haploid genotypes present (see Allele_Frequency.hh ); many
commands treat these as missing.
查看结果,结果文件是Allele_Frequency.frq
$ head Allele_Frequency.frq
CHR SNP A1 A2 MAF NCHROBS
1 rs12565286 C G 0.0678 118
1 rs12138618 A G 0.05833 120
1 rs3094315 G A 0.1552 116
1 rs3131968 A G 0.125 120
1 rs12562034 A G 0.09167 120
1 rs2905035 A G 0.125 120
1 rs12124819 G A 0.3417 120
1 rs2980319 A T 0.125 120
1 rs4040617 G A 0.125 120
如果对其进行质控,用–maf 0.01,会去掉maf小于0.01的位点。
6.2 缺失
缺失包括样本缺失率统计和位点缺失率统计。
命令:
plink --bfile hapmap-ceu --missing --out missing_data
日志:
$ plink --bfile hapmap-ceu --missing --out missing_data
PLINK v1.90b6.21 64-bit (19 Oct 2020) www.cog-genomics.org/plink/1.9/
(C) 2005-2020 Shaun Purcell, Christopher Chang GNU General Public License v3
Logging to missing_data.log.
Options in effect:
--bfile hapmap-ceu
--missing
--out missing_data
15236 MB RAM detected; reserving 7618 MB for main workspace.
2239392 variants loaded from .bim file.
60 people (30 males, 30 females) loaded from .fam.
Using 1 thread (no multithreaded calculations invoked).
Before main variant filters, 60 founders and 0 nonfounders present.
Calculating allele frequencies... done.
Total genotyping rate is 0.992022.
--missing: Sample missing data report written to missing_data.imiss, and
variant-based missing data report written to missing_data.lmiss.
Warning: 59 het. haploid genotypes present (see missing_data.hh ); many
commands treat these as missing.
查看样本缺失的文件:
$ head missing_data.imiss
FID IID MISS_PHENO N_MISS N_GENO F_MISS
1334 NA12144 Y 15077 2239392 0.006733
1334 NA12145 Y 19791 2239392 0.008838
1334 NA12146 Y 13981 2239392 0.006243
1334 NA12239 Y 14072 2239392 0.006284
1340 NA06994 Y 16080 2239392 0.007181
1340 NA07000 Y 26113 2239392 0.01166
1340 NA07022 Y 17467 2239392 0.0078
1340 NA07056 Y 12133 2239392 0.005418
1341 NA07034 Y 20425 2239392 0.009121
查看位点缺失的文件:
$ head missing_data.lmiss
CHR SNP N_MISS N_GENO F_MISS
1 rs12565286 1 60 0.01667
1 rs12138618 0 60 0
1 rs3094315 2 60 0.03333
1 rs3131968 0 60 0
1 rs12562034 0 60 0
1 rs2905035 0 60 0
1 rs12124819 0 60 0
1 rs2980319 0 60 0
1 rs4040617 0 60 0
6.3 哈温检测
–hardy
代码:
plink --bfile 1kg_EU_qc --hardy
日志:
PLINK v1.90b5.3 64-bit (21 Feb 2018) www.cog-genomics.org/plink/1.9/
(C) 2005-2018 Shaun Purcell, Christopher Chang GNU General Public License v3
Logging to plink.log.
Options in effect:
--bfile 1kg_EU_qc
--hardy
1031523 MB RAM detected; reserving 515761 MB for main workspace.
851065 variants loaded from .bim file.
379 people (178 males, 201 females) loaded from .fam.
Using 1 thread (no multithreaded calculations invoked).
Before main variant filters, 379 founders and 0 nonfounders present.
Calculating allele frequencies... done.
--hardy: Writing Hardy-Weinberg report (founders only) to plink.hwe ... done.
结果:
[dengfei@localhost 08_chapter]$ head plink.hwe
CHR SNP TEST A1 A2 GENO O(HET) E(HET) P
1 rs1048488 ALL(NP) C T 9/136/234 0.3588 0.3238 0.03894
1 rs3115850 ALL(NP) T C 8/135/236 0.3562 0.319 0.02412
1 rs2519031 ALL(NP) G A 0/11/368 0.02902 0.0286 1
1 rs4970383 ALL(NP) A C 22/149/208 0.3931 0.3796 0.5881
1 rs4475691 ALL(NP) T C 13/118/248 0.3113 0.3078 1
1 rs1806509 ALL(NP) C A 67/171/141 0.4512 0.4809 0.2402
1 rs7537756 ALL(NP) G A 14/124/241 0.3272 0.3206 0.8725
1 rs28576697 ALL(NP) C T 37/161/181 0.4248 0.4278 0.9045
1 rs7523549 ALL(NP) T C 0/28/351 0.07388 0.07115 1
[dengfei@localhost 08_chapter]$ wc -l plink.hwe
851066 plink.hwe
6.4 杂合率检测
–het
代码:
plink --bfile 1kg_EU_qc --het
日志:
PLINK v1.90b5.3 64-bit (21 Feb 2018) www.cog-genomics.org/plink/1.9/
(C) 2005-2018 Shaun Purcell, Christopher Chang GNU General Public License v3
Logging to plink.log.
Options in effect:
--bfile 1kg_EU_qc
--het
1031523 MB RAM detected; reserving 515761 MB for main workspace.
851065 variants loaded from .bim file.
379 people (178 males, 201 females) loaded from .fam.
Using 1 thread (no multithreaded calculations invoked).
Before main variant filters, 379 founders and 0 nonfounders present.
Calculating allele frequencies... done.
851065 variants and 379 people pass filters and QC.
Note: No phenotypes present.
--het: 851065 variants scanned, report written to plink.het .
结果:
[dengfei@localhost 08_chapter]$ head plink.het
FID IID O(HOM) E(HOM) N(NM) F
0 HG00096 582990 5.838e+05 851065 -0.003127
0 HG00097 582246 5.838e+05 851065 -0.005911
0 HG00099 582456 5.838e+05 851065 -0.005125
0 HG00100 582700 5.838e+05 851065 -0.004212
0 HG00101 581527 5.838e+05 851065 -0.008602
0 HG00102 585010 5.838e+05 851065 0.004432
0 HG00103 583984 5.838e+05 851065 0.0005925
0 HG00104 586189 5.838e+05 851065 0.008844
0 HG00106 583868 5.838e+05 851065 0.0001584
[dengfei@localhost 08_chapter]$ wc -l plink.het
380 plink.het
7. 数据质控qc
7.1 样本质控–mind
代码:
plink --bfile 1kg_EU_qc --mind 0.1 --recode --out re1
日志:
PLINK v1.90b5.3 64-bit (21 Feb 2018) www.cog-genomics.org/plink/1.9/
(C) 2005-2018 Shaun Purcell, Christopher Chang GNU General Public License v3
Logging to re1.log.
Options in effect:
--bfile 1kg_EU_qc
--mind 0.1
--out re1
--recode
1031523 MB RAM detected; reserving 515761 MB for main workspace.
851065 variants loaded from .bim file.
379 people (178 males, 201 females) loaded from .fam.
0 people removed due to missing genotype data (--mind).
Using 1 thread (no multithreaded calculations invoked).
Before main variant filters, 379 founders and 0 nonfounders present.
Calculating allele frequencies... done.
851065 variants and 379 people pass filters and QC.
Note: No phenotypes present.
--recode ped to re1.ped + re1.map ... done.
结果:
[dengfei@localhost 08_chapter]$ wc -l re1.*
24 re1.log
851065 re1.map
379 re1.ped
851468 总用量
7.2 位点质控–geno
代码:
plink --bfile 1kg_EU_qc --geno 0.1 --recode --out re1
日志:
PLINK v1.90b5.3 64-bit (21 Feb 2018) www.cog-genomics.org/plink/1.9/
(C) 2005-2018 Shaun Purcell, Christopher Chang GNU General Public License v3
Logging to re1.log.
Options in effect:
--bfile 1kg_EU_qc
--geno 0.1
--out re1
--recode
1031523 MB RAM detected; reserving 515761 MB for main workspace.
851065 variants loaded from .bim file.
379 people (178 males, 201 females) loaded from .fam.
Using 1 thread (no multithreaded calculations invoked).
Before main variant filters, 379 founders and 0 nonfounders present.
Calculating allele frequencies... done.
0 variants removed due to missing genotype data (--geno).
851065 variants and 379 people pass filters and QC.
Note: No phenotypes present.
--recode ped to re1.ped + re1.map ... done.
结果:
[dengfei@localhost 08_chapter]$ wc -l re1*
24 re1.log
851065 re1.map
379 re1.ped
851468 总用量
7.3 maf质控–maf
代码:
plink --bfile 1kg_EU_qc --maf 0.01 --recode --out re1
日志:
PLINK v1.90b5.3 64-bit (21 Feb 2018) www.cog-genomics.org/plink/1.9/
(C) 2005-2018 Shaun Purcell, Christopher Chang GNU General Public License v3
Logging to re1.log.
Options in effect:
--bfile 1kg_EU_qc
--maf 0.01
--out re1
--recode
1031523 MB RAM detected; reserving 515761 MB for main workspace.
851065 variants loaded from .bim file.
379 people (178 males, 201 females) loaded from .fam.
Using 1 thread (no multithreaded calculations invoked).
Before main variant filters, 379 founders and 0 nonfounders present.
Calculating allele frequencies... done.
17767 variants removed due to minor allele threshold(s)
(--maf/--max-maf/--mac/--max-mac).
833298 variants and 379 people pass filters and QC.
Note: No phenotypes present.
--recode ped to re1.ped + re1.map ... done.
结果:
[dengfei@localhost 08_chapter]$ wc -l re1*
25 re1.log
833298 re1.map
379 re1.ped
833702 总用量
7.4 哈温质控 --hwe
代码:
plink --bfile 1kg_EU_qc --hwe 1e-5 --recode --out re1
日志:
PLINK v1.90b5.3 64-bit (21 Feb 2018) www.cog-genomics.org/plink/1.9/
(C) 2005-2018 Shaun Purcell, Christopher Chang GNU General Public License v3
Logging to re1.log.
Options in effect:
--bfile 1kg_EU_qc
--hwe 1e-5
--out re1
--recode
1031523 MB RAM detected; reserving 515761 MB for main workspace.
851065 variants loaded from .bim file.
379 people (178 males, 201 females) loaded from .fam.
Using 1 thread (no multithreaded calculations invoked).
Before main variant filters, 379 founders and 0 nonfounders present.
Calculating allele frequencies... done.
--hwe: 25 variants removed due to Hardy-Weinberg exact test.
851040 variants and 379 people pass filters and QC.
Note: No phenotypes present.
--recode ped to re1.ped + re1.map ... done.
结果:
[dengfei@localhost 08_chapter]$ wc -l re1*
24 re1.log
851040 re1.map
379 re1.ped
851443 总用量