Title
题目
Cross-modality deep feature learning for brain tumor segmentation
用于脑肿瘤分割的跨模态深度特征学习
01
文献速递介绍
作为最致命的流行病,脑肿瘤的研究越来越受到关注。本文研究了一种基于深度学习的自动分割胶质瘤的方法,称为脑肿瘤分割(BTS)。在这个任务中,医学图像包含四种MRI模态,分别是T1加权(T1)模态、增强T1加权(T1c)模态、T2加权(T2)模态和液体衰减逆转(FLAIR)模态。目标是分割三个不同的目标区域,分别是整个肿瘤区域、肿瘤核心区域和增强肿瘤核心区域。多模态数据及相应的肿瘤区域标签示例如图1所示。
随着深度学习技术的快速发展,深度卷积神经网络(DCNNs)已被引入医学图像分析领域,并广泛应用于脑肿瘤分割。鉴于已建立的DCNN模型,现有的脑肿瘤分割方法通常将此任务视为一个多类别像素级分类问题,就像普通RGB图像数据上的语义分割任务一样。然而,通过忽略医学图像数据与普通RGB图像数据之间的巨大差异,这种方法不会获得最佳解决方案。具体来说,这两种数据之间存在着两个明显的不同特性:1)非常大规模的RGB图像数据可以通过智能手机或相机从我们的日常生活中获取。然而,医学图像数据非常稀缺,尤其是对应的需要专业知识且往往非常耗时的手动标注。2)与普通RGB图像数据不同,医学图像数据(对于所研究的脑肿瘤分割任务和其他任务)通常由捕获不同病理特性的多个MRI模态组成。
由于上述特点,脑肿瘤分割仍然存在需要解决的挑战性问题。具体而言,由于数据规模不足,训练DCNN模型可能会遭受过拟合问题,因为DCNN模型通常包含大量网络参数。这增加了训练适用于脑肿瘤分割的理想DCNN模型的难度。其次,由于复杂的数据结构,直接连接多模态数据以形成网络输入,就像以前的工作中所做的那样,既不是充分利用每个模态数据背后知识的最佳选择,也不是融合来自多模态数据知识的有效策略。
Abstract
摘要
Recent advances in machine learning and prevalence of digital medical images have opened up an opportunity to address the challenging brain tumor segmentation (BTS) task by using deep convolutional neuralnetworks. However, different from the RGB image data that are very widespread, the medical image dataused in brain tumor segmentation are relatively scarce in terms of the data scale but contain the richerinformation in terms of the modality property. To this end, this paper proposes a novel cross-modalitydeep feature learning framework to segment brain tumors from the multi-modality MRI data. The coreidea is to mine rich patterns across the multi-modality data to make up for the insufficient data scale.The proposed cross-modality deep feature learning framework consists of two learning processes: thecross-modality feature transition (CMFT) process and the cross-modality feature fusion (CMFF) process,which aims at learning rich feature representations by transiting knowledge across different modalitydata and fusing knowledge from different modality data, respectively. Comprehensive experiments areconducted on the BraTS benchmarks, which show that the proposed cross-modality deep feature learning framework can effectively improve the brain tumor
最近机器学习的进展和数字医学图像的普及为利用深度卷积神经网络解决具有挑战性的脑肿瘤分割(BTS)任务开辟了机遇。然而,与非常普遍的RGB图像数据不同,用于脑肿瘤分割的医学图像数据在数据规模方面相对较少,但在模态属性方面包含了更丰富的信息。因此,本文提出了一种新颖的跨模态深度特征学习框架,以从多模态MRI数据中分割脑肿瘤。其核心思想是挖掘跨模态数据中的丰富模式,弥补数据规模不足的问题。所提出的跨模态深度特征学习框架包括两个学习过程:跨模态特征转换(CMFT)过程和跨模态特征融合(CMFF)过程,分别旨在通过在不同模态数据之间转移知识和融合来自不同模态数据的知识来学习丰富的特征表示。在BraTS基准测试上进行了全面实验,结果表明所提出的跨模态深度特征学习框架能够有效改善脑肿瘤分割。
Conclusion
结论
In this work, we have proposed a novel cross-modality deepfeature learning framework for segmenting brain tumor areas fromthe multi-modality MR scans. Considering that the medical imagedata for brain tumor segmentation are relatively scarce in termsof the data scale but containing the richer information in terms ofthe modality property, we propose to mine rich patterns across themulti-modality data to make up for the insufficiency in data scale.The proposed learning framework consists of a cross-modality feature transition (CMFT) process and a cross-modality feature fusion(CMFF) process. By building a generative adversarial network-basedlearning scheme to implement the cross-modality feature transitionprocess, our approach is able to learn useful feature representationsfrom the knowledge transition across different modality data withoutany human annotation. While the cross-modality feature fusion process transfers the features learned from the feature transition processand is empowered with the novel fusion branch to guide a strong fea-ture fusion process. Comprehensive experiments are conducted ontwo BraTS benchmarks, which demonstrate the effectiveness of ourapproach when compared to baseline models and state-of-the-artmethods. *To our knowledge, one limitation of this work is the current learning framework requires that the network architectures of themodal generator and the segmentation predictor be almost the same.To address this inconvenience, one potential future direction is to introduce the knowledge distillation mechanism to replace thesimple parameter transfer process.
在这项工作中,我们提出了一种新颖的跨模态深度特征学习框架,用于从多模态MR扫描中分割脑肿瘤区域。考虑到用于脑肿瘤分割的医学图像数据在数据规模方面相对稀缺,但在模态属性方面包含了更丰富的信息,我们提出了在多模态数据中挖掘丰富模式的方法来弥补数据规模不足的问题。所提出的学习框架包括跨模态特征转换(CMFT)过程和跨模态特征融合(CMFF)过程。通过建立一个基于生成对抗网络的学习方案来实现跨模态特征转换过程,我们的方法能够从不同模态数据之间的知识转移中学习到有用的特征表示,而无需任何人工注释。而跨模态特征融合过程将从特征转换过程学到的特征进行转移,并利用新颖的融合分支来指导强大的特征融合过程。我们在两个BraTS基准测试上进行了全面实验,结果表明与基线模型和最新方法相比,我们的方法的有效性。据我们所知,这项工作的一个局限是当前的学习框架要求模态生成器和分割预测器的网络架构几乎相同。为了解决这个不便,一个潜在的未来方向是引入知识蒸馏机制来替代简单的参数传递过程。
Figure
图
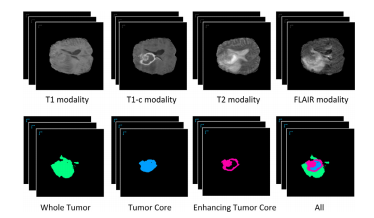
Fig. 1. An illustration of the brain tumor segmentation task. The top four volumedata are the multi-modality MR image data. The segmentation labels for the WholeTumor area (WT), Tumor Core area (TC), Enhancing Tumor Core area (WT), and alltypes of tumor areas are shown in the bottom row. The regions without coloredmasks are normal areas.
图1. 脑肿瘤分割任务的示意图。顶部的四个体积数据是多模态MR图像数据。底部一行显示了整个肿瘤区域(WT)、肿瘤核心区域(TC)、增强肿瘤核心区域(ET)和所有类型的肿瘤区域的分割标签。没有彩色掩膜的区域是正常区域。
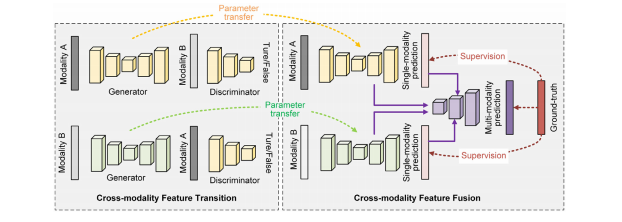
Fig. 2. An illustration of the proposed cross-modality deep feature learning framework for brain tumor segmentation. To be brief and to the point, we only show the learningframework by using two-modality data.
图2. 所提出的用于脑肿瘤分割的跨模态深度特征学习框架的示意图。为简洁明了起见,我们仅展示了使用两种模态数据的学习框架。
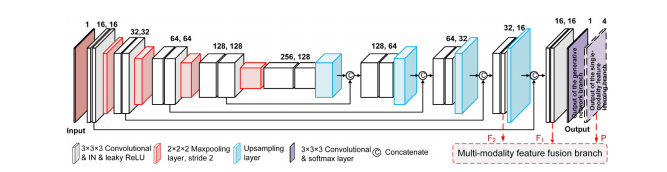
Fig. 3. Illustration of the detailed architecture of the generator, where IN is short for instance normalization. Notice that this is also the architecture of the single-modalityfeature learning branch. The only difference between these two network branches is the last output layer, where the output of the generator is drawn in the solid linewhile the output of the single-modality feature learning branch is drawn in the dashed line. The deep features in the last two convolutional layers, as well as the output ofthe single-modality feature learning branch, are connected to the cross-modality feature fusion branch, which is annotated in red. For ease of understanding, we show thenetwork in 2D convolution-like architecture. While we actually use the 3D convolution in network layers.
图3. 生成器的详细架构示意图,其中IN代表实例标准化。请注意,这也是单模态特征学习分支的架构。这两个网络分支之间唯一的区别在于最后的输出层,其中生成器的输出用实线表示,而单模态特征学习分支的输出用虚线表示。最后两个卷积层的深度特征,以及单模态特征学习分支的输出,与跨模态特征融合分支相连接,用红色标注。为了便于理解,我们展示了类似于2D卷积的网络架构。而实际上我们在网络层中使用了3D卷积。
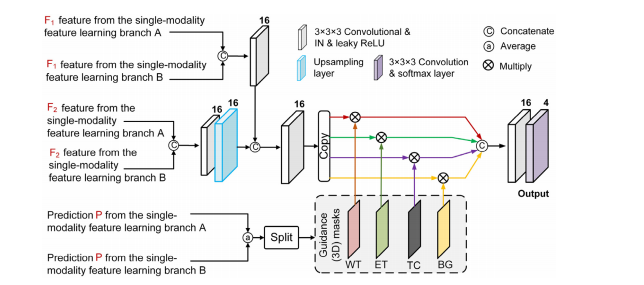
Fig. 4. Illustration of the cross-modality feature fusion branch with the mask-guided feature learning scheme, where IN is short for instance normalization. For ease ofunderstanding, we show the network in 2D convolution-like architecture. While we actually use the 3D convolution in network layers.
图4. 使用掩模引导特征学习方案的跨模态特征融合分支的示意图,其中IN代表实例标准化。为了便于理解,我们展示了类似于2D卷积的网络架构。而实际上我们在网络层中使用了3D卷积。
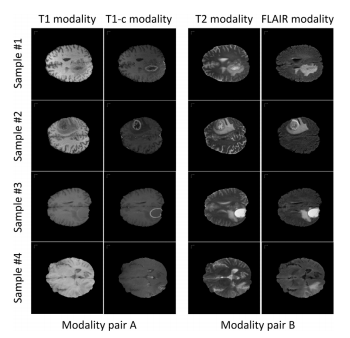
Fig. 5. Examples of the modality pairs, where we use the T1 modality and T1cmodality to form the modality pair A while using the T2 modality and FLAIR modality to form the modality pair B. From the examples we can observe that the information contained within each modality pair is relatively consistent while the information contained across the different modality pairs is relatively distinct and complementary. This enables the cross-modality feature transition process to learn richpatterns.
图5. 模态对的示例,我们使用T1模态和T1c模态形成模态对A,同时使用T2模态和FLAIR模态形成模态对B。从示例中我们可以观察到,每个模态对内包含的信息相对一致,而在不同模态对之间包含的信息相对独特且互补。这使得跨模态特征转换过程能够学习到丰富的模式。
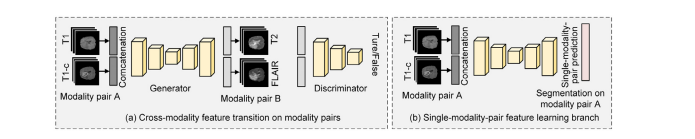
Fig. 6. Illustration of the proposed strategy to extend the proposed cross-modality deep feature learning framework to work on modality quaternions. In the cross-modalityfeature transition process, we convert the input and output from one modality data to the concatenation of an modality pair. While in the cross-modality feature fusionprocess, we convert the single-modality feature learning branch to the single-modality-pair feature learning branch, which predicts the segmentation masks of each singlemodality-pair.
图6. 展示了将所提出的跨模态深度特征学习框架扩展到模态四元数上的策略。在跨模态特征转换过程中,我们将输入和输出从一个模态数据转换为模态对的连接。而在跨模态特征融合过程中,我们将单模态特征学习分支转换为单模态对特征学习分支,该分支预测每个单模态对的分割掩膜。
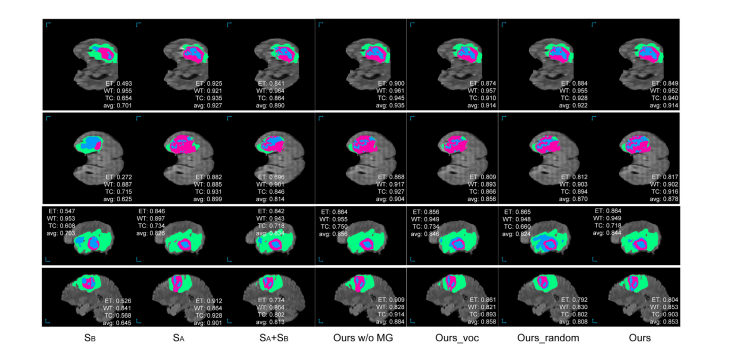
Fig. 7. Examples of the segmentation results of the proposed approach as well as the compared baseline methods on the BraTS 2018 validation set. As the ground-truth segmentation annotation is not acquirable, we annotate the dice score for the segmented tumor regions on each test sample instead of showing the ground-truth segmentationannotation. The WT, TC, and ET areas are masked in green, blue, and purple, respectively.
图7. 所提出的方法以及与之比较的基线方法在BraTS 2018验证集上的分割结果示例。由于无法获取地面真实的分割注释,我们在每个测试样本上注释了分割肿瘤区域的Dice分数,而不是显示地面真实的分割注释。WT、TC和ET区域分别用绿色、蓝色和紫色标记。

Fig. 8. Comparison of the segmentation results and the ground-truth annotation on the BraTS 2018 training set. Notice that the average Dice score on the BraTS 2018 trainingset is 0.886, which is moderately higher than the Dice score on the BraTS 2018 validation set. The WT, TC, and ET areas are masked in green, blue, and purple, respectively.
图8. 在BraTS 2018训练集上比较分割结果和地面真实标注。请注意,BraTS 2018训练集上的平均Dice分数为0.886,略高于BraTS 2018验证集上的Dice分数。WT、TC和ET区域分别用绿色、蓝色和紫色标记。
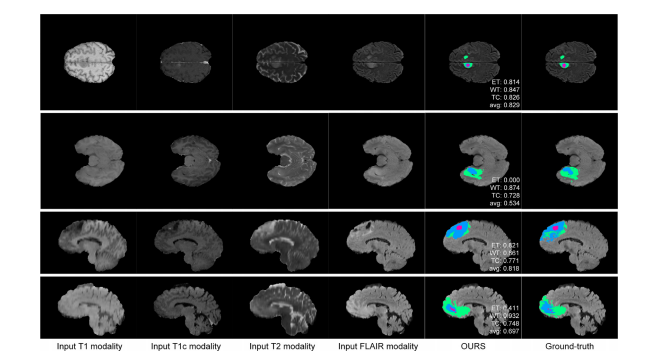
Fig. 9. Examples of the failure cases on the BraTS 2018 training set, where the WT, TC, and ET areas are masked in green, blue, and purple, respectively. The first exampleis a failure case in the HGG subjects, which is mainly due to the inaccurate tumor boundaries. The other examples are from the LGG cases, where the absent ET area,discontinuous tumor regions and ragged tumor contours make the model hard to predict.
图9. 在BraTS 2018训练集上的失败案例示例,其中WT、TC和ET区域分别用绿色、蓝色和紫色标记。第一个示例是HGG受试者的失败案例,主要是由于肿瘤边界不准确。其他示例来自LGG病例,其中缺失的ET区域、不连续的肿瘤区域和参差不齐的肿瘤轮廓使得模型难以预测。
Table
表
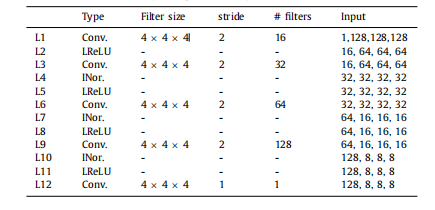
Table 1The architecture of the discriminator network branch. In the “Input” block, the firstdimension is the number of channels and the next three dimensions are the sizeof the feature maps. Conv. is short for the 3D convolution, and # filters indicatesthe number of filters. Notice that when learning on modality quaternions mentioned**in Section 3.3, the number of the input channel of L1 becomes 2
表1:鉴别器网络分支的架构。在“输入”块中,第一个维度是通道数,接下来的三个维度是特征图的大小。Conv.表示3D卷积,# filters表示滤波器的数量。请注意,当学习模态四元数时,第3.3节中提到的L1的输入通道数变为2
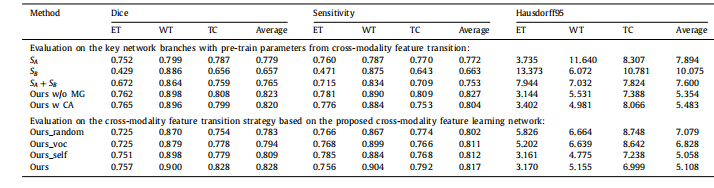
Table 2Comparison results of the proposed approach and the other baseline models on the BraTS 2017 validation set. Higher Dice and Sensitivity scores indicate the better results,while lower Hausdorff95 scores indicate the better results.
表2:在BraTS 2017验证集上,所提出的方法与其他基线模型的比较结果。较高的Dice和灵敏度分数表示较好的结果,而较低的Hausdorff95分数表示较好的结果。
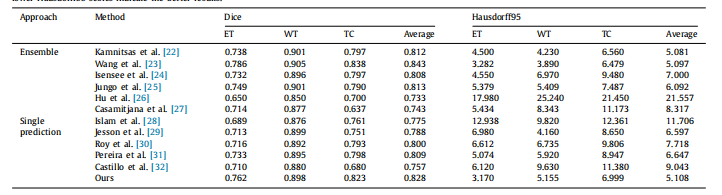
Table 3Comparison results of the proposed approach and the other state-of-the-art models on the BraTS 2017 validation set. Higher Dice scores indicate the better results, whilelower Hausdorff95 scores indicate the better results.
表3:在BraTS 2017验证集上,所提出的方法与其他最新模型的比较结果。较高的Dice分数表示较好的结果,而较低的Hausdorff95分数表示较好的结果。
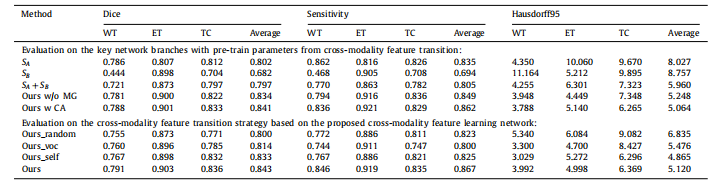
Table 4Comparison results of the proposed approach and the other baseline models on the BraTS 2018 validation set. Higher Dice and Sensitivity scores indicate the better results,while lower Hausdorff95 scores indicate the better results.
表4:在BraTS 2018验证集上,所提出的方法与其他基线模型的比较结果。较高的Dice和灵敏度分数表示较好的结果,而较低的Hausdorff95分数表示较好的结果。
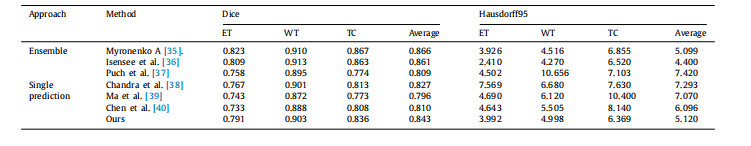
Table 5Comparison results of the proposed approach and the other state-of-the-art models on the BraTS 2018 validation set. Higher Dice scores indicate the better results, whilelower Hausdorff95 scores indicate the better results
表5:在BraTS 2018验证集上,所提出的方法与其他最新模型的比较结果。较高的Dice分数表示较好的结果,而较低的Hausdorff95分数表示较好的结果。