欢迎关注我的CSDN:https://spike.blog.csdn.net/
本文地址:https://spike.blog.csdn.net/article/details/134595717
GPCRs(G Protein-Coupled Receptors,G蛋白偶联受体),又称为7次跨膜受体,是细胞信号传导中的重要蛋白质,当膜外的配体作用于该受体时,该受体的膜内部分与G蛋白相互结合激活G蛋白,进而启动不同的信号转导通路,参与多种生理和病理过程,如免疫调节、行为和情绪的调节、感觉的传递、内稳态的调节以及肿瘤的生长与转移等。G 蛋白偶联受体(GPCR)超家族包含约 600-1000个靶标,是已知最大的一类具有治疗价值的分子靶标,目前世界药物市场上至少有三分之一的小分子药物是GPCR的激动剂或拮抗剂。GPCR过表达细胞系已广泛用于药物筛选和自身免疫性疾病的病因和发病机制研究。
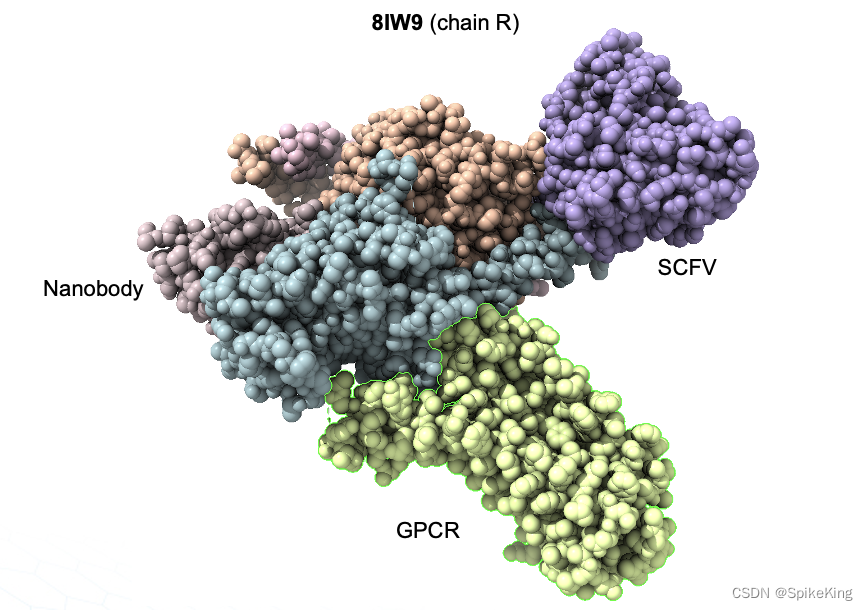
1. GPCR 数据集
GPCR 数据,来自于 GPCR-DB 官网,其中包括 PDB、GPCR chain id、PDB Date 等信息,再从 PDB 中获取 FASTA Sequence 信息,即:
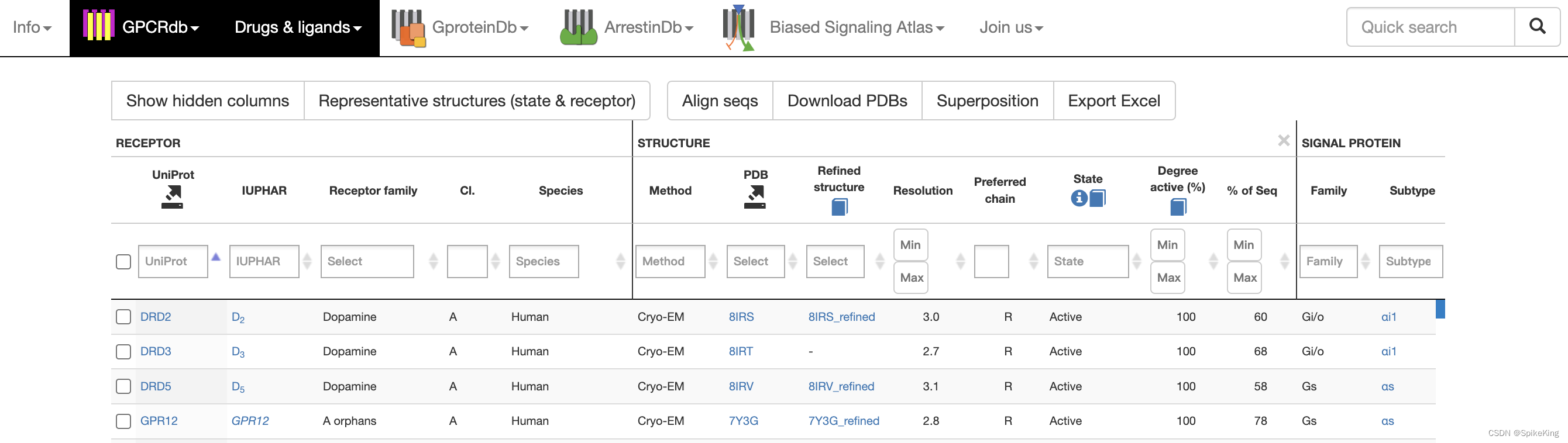
PDB 的 FASTA Sequence,来自于 RCSB 官网,如下:
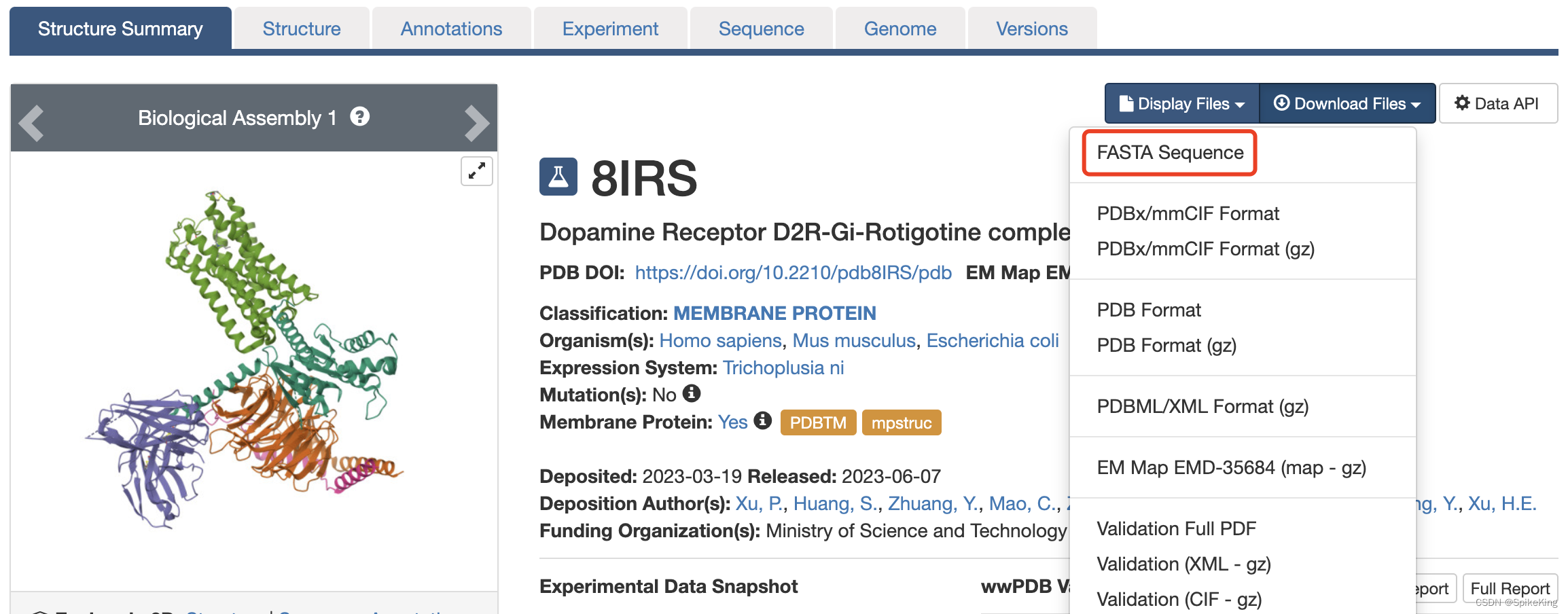
8IRS 的 FASTA 序列,如下:
>8IRS_1|Chain A|Guanine nucleotide-binding protein G(i) subunit alpha-1|Homo sapiens (9606)
MGCTLSAEDKAAVERSKMIDRNLREDGEKAAREVKLLLLGAGESGKNTIVKQMKIIHEAGYSEEECKQYKAVVYSNTIQSIIAIIRAMGRLKIDFGDSARADDARQLFVLAGAAEEGFMTAELAGVIKRLWKDSGVQACFNRSREYQLNDSAAYYLNDLDRIAQPNYIPTQQDVLRTRVKTTGIVETHFTFKDLHFKMFDVGAQRSERKKWIHCFEGVTAIIFCVALSDYDLVLAEDEEMNRMHASMKLFDSICNNKWFTDTSIILFLNKKDLFEEKIKKSPLTICYPEYAGSNTYEEAAAYIQCQFEDLNKRKDTKEIYTHFTCSTDTKNVQFVFDAVTDVIIKNNLKDCGLF
>8IRS_2|Chain B|Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1|Homo sapiens (9606)
MGSLLQSELDQLRQEAEQLKNQIRDARKACADATLSQITNNIDPVGRIQMRTRRTLRGHLAKIYAMHWGTDSRLLVSASQDGKLIIWDSYTTNKVHAIPLRSSWVMTCAYAPSGNYVACGGLDNICSIYNLKTREGNVRVSRELAGHTGYLSCCRFLDDNQIVTSSGDTTCALWDIETGQQTTTFTGHTGDVMSLSLAPDTRLFVSGACDASAKLWDVREGMCRQTFTGHESDINAICFFPNGNAFATGSDDATCRLFDLRADQELMTYSHDNIICGITSVSFSKSGRLLLAGYDDFNCNVWDALKADRAGVLAGHDNRVSCLGVTDDGMAVATGSWDSFLKIWN
>8IRS_3|Chain C[auth E]|ScFv16|Mus musculus (10090)
DVQLVESGGGLVQPGGSRKLSCSASGFAFSSFGMHWVRQAPEKGLEWVAYISSGSGTIYYADTVKGRFTISRDDPKNTLFLQMTSLRSEDTAMYYCVRSIYYYGSSPFDFWGQGTTLTVSSGGGGSGGGGSGGGGSDIVMTQATSSVPVTPGESVSISCRSSKSLLHSNGNTYLYWFLQRPGQSPQLLIYRMSNLASGVPDRFSGSGSGTAFTLTISRLEAEDVGVYYCMQHLEYPLTFGAGTKLELK
>8IRS_4|Chain D[auth G]|Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2|Homo sapiens (9606)
MASNNTASIAQARKLVEQLKMEANIDRIKVSKAAADLMAYCEAHAKEDPLLTPVPASENPFREKKFFCAIL
>8IRS_5|Chain E[auth R]|Soluble cytochrome b562,D(2) dopamine receptor|Escherichia coli (562)
DYKDDDDAKLQTMHHHHHHHHHHHHHHHADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLASENLYFQGGTMDPLNLSWYDDDLERQNWSRPFNGSDGKADRPHYNYYATLLTLLIAVIVFGNVLVCMAVSREKALQTTTNYLIVSLAVADLLVATLVMPWVVYLEVVGEWKFSRIHCDIFVTLDVMMCTASILNLCAISIDRYTAVAMPMLYNTRYSSKRRVTVMISIVWVLSFTISCPLLFGLNNADQNECIIANPAFVVYSSIVSFYVPFIVTLLVYIKIYIVLRRRRKRVNTKRSSRAFRAHLRAPLKGNCTHPEDMKLCTVIMKSNGSFPVNRRRVEAARRAQELEMEMLSSTSPPERTRYSPIPPSHHQLTLPDPSHHGLHSTPDSPAKPEKNGHAKDHPKIAKIFEIQTMPNGKTRTSLKTMSRRKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCNIPPVLYSAFTWLGYVNSAVNPIIYTTFNIEFRKAFLKILHC
存储的 8irs 序列,即:
pdb,chain,seq,mol,gpcr_chain
8irs,"R,G,E,B,A","DYKDDDDAKLQTMHHHHHHHHHHHHHHHADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLASENLYFQGGTMDPLNLSWYDDDLERQNWSRPFNGSDGKADRPHYNYYATLLTLLIAVIVFGNVLVCMAVSREKALQTTTNYLIVSLAVADLLVATLVMPWVVYLEVVGEWKFSRIHCDIFVTLDVMMCTASILNLCAISIDRYTAVAMPMLYNTRYSSKRRVTVMISIVWVLSFTISCPLLFGLNNADQNECIIANPAFVVYSSIVSFYVPFIVTLLVYIKIYIVLRRRRKRVNTKRSSRAFRAHLRAPLKGNCTHPEDMKLCTVIMKSNGSFPVNRRRVEAARRAQELEMEMLSSTSPPERTRYSPIPPSHHQLTLPDPSHHGLHSTPDSPAKPEKNGHAKDHPKIAKIFEIQTMPNGKTRTSLKTMSRRKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCNIPPVLYSAFTWLGYVNSAVNPIIYTTFNIEFRKAFLKILHC,MASNNTASIAQARKLVEQLKMEANIDRIKVSKAAADLMAYCEAHAKEDPLLTPVPASENPFREKKFFCAIL,DVQLVESGGGLVQPGGSRKLSCSASGFAFSSFGMHWVRQAPEKGLEWVAYISSGSGTIYYADTVKGRFTISRDDPKNTLFLQMTSLRSEDTAMYYCVRSIYYYGSSPFDFWGQGTTLTVSSGGGGSGGGGSGGGGSDIVMTQATSSVPVTPGESVSISCRSSKSLLHSNGNTYLYWFLQRPGQSPQLLIYRMSNLASGVPDRFSGSGSGTAFTLTISRLEAEDVGVYYCMQHLEYPLTFGAGTKLELK,MGSLLQSELDQLRQEAEQLKNQIRDARKACADATLSQITNNIDPVGRIQMRTRRTLRGHLAKIYAMHWGTDSRLLVSASQDGKLIIWDSYTTNKVHAIPLRSSWVMTCAYAPSGNYVACGGLDNICSIYNLKTREGNVRVSRELAGHTGYLSCCRFLDDNQIVTSSGDTTCALWDIETGQQTTTFTGHTGDVMSLSLAPDTRLFVSGACDASAKLWDVREGMCRQTFTGHESDINAICFFPNGNAFATGSDDATCRLFDLRADQELMTYSHDNIICGITSVSFSKSGRLLLAGYDDFNCNVWDALKADRAGVLAGHDNRVSCLGVTDDGMAVATGSWDSFLKIWN,MGCTLSAEDKAAVERSKMIDRNLREDGEKAAREVKLLLLGAGESGKNTIVKQMKIIHEAGYSEEECKQYKAVVYSNTIQSIIAIIRAMGRLKIDFGDSARADDARQLFVLAGAAEEGFMTAELAGVIKRLWKDSGVQACFNRSREYQLNDSAAYYLNDLDRIAQPNYIPTQQDVLRTRVKTTGIVETHFTFKDLHFKMFDVGAQRSERKKWIHCFEGVTAIIFCVALSDYDLVLAEDEEMNRMHASMKLFDSICNNKWFTDTSIILFLNKKDLFEEKIKKSPLTICYPEYAGSNTYEEAAAYIQCQFEDLNKRKDTKEIYTHFTCSTDTKNVQFVFDAVTDVIIKNNLKDCGLF",R
注意:Chain ID 优先使用 auth 值,其次使用默认的 Chain 值,例如 Chain D[auth G] 是 G,Chain A 是 A。
获取 CSV 文件 gpcr_dataset.csv,字段包括:pdb、chain、seq、gpcr_chain 等关键字段。
2. 导出 PDB 与 FASTA 数据
根据数据文件,导出 GPCR 相关的复合物 PDB 与 多链 FASTA,再根据 GPCR 链 (Preferred Chain) 导出单体 PDB 与 单链 FASTA。
2.1 导出 PDB 复合物与蛋白质序列
调用 p1_main_gpcr_pdb_exporter.py,提取 GPCR 复合物结构与序列。
python3 gpcr/p1_main_gpcr_pdb_exporter.py \
-i gpcr/gpcr_data_filter.csv \
-p gpcr/gpcr_complex_v2/pdb_complex_446 \
-f gpcr/gpcr_complex_v2/fasta_complex_446
存储的 PDB 的格式与 FASTA 的格式,建议保留 PDB 名称、链名、链长、全部序列长度,即:
- PDB:
8IW9_A234_B338_C53_R309_S231_N128_1293.pdb - FASTA:
8IW9_A362_B377_C59_N128_R348_S285_1559.fasta
注意:全长的 FASTA 序列预测 PDB 结构,优于从 PDB 中提取的 FASTA 序列预测的结构,原因是 PDB 结构中缺失一些残基位置。
GPCR 复合物的输出位置,包括 PDB 与 FASTA,即:
gpcr/gpcr_complex_v2/pdb_complex_446/
gpcr/gpcr_complex_v2/fasta_complex_446/
8IW9_A362_B377_C59_N128_R348_S285_1559.fasta 序列:
>A
MMGCTLSAEDKAAVERSKMIEKQLQKDKQVYRATHRLLLLGADNSGKSTIVKQMRIYHVNGYSEEECKQYKAVVYSNTIQSIIAIIRAMGRLKIDFGDSARADDARQLFVLAGAAEEGFMTAELAGVIKRLWKDSGVQACFNRSREYQLNDSAAYYLNDLDRIAQPNYIPTQQDVLRTRVKTSGIFETKFQVDKVNFHMFDVGAQRDERRKWIQCFNDVTAIIFVVDSSDYNRLQEALNDFKSIWNNRWLRTISVILFLNKQDLLAEKVLAGKSKIEDYFPEFARYTTPEDATPEPGEDPRVTRAKYFIRDEFLRISTASGDGRHYCYPHFTCSVDTENARRIFNDCRDIIQRMHLRQYELL
>B
MHHHHHHGSLLQSELDQLRQEAEQLKNQIRDARKACADATLSQITNNIDPVGRIQMRTRRTLRGHLAKIYAMHWGTDSRLLVSASQDGKLIIWDSYTTNKVHAIPLRSSWVMTCAYAPSGNYVACGGLDNICSIYNLKTREGNVRVSRELAGHTGYLSCCRFLDDNQIVTSSGDTTCALWDIETGQQTTTFTGHTGDVMSLSLAPDTRLFVSGACDASAKLWDVREGMCRQTFTGHESDINAICFFPNGNAFATGSDDATCRLFDLRADQELMTYSHDNIICGITSVSFSKSGRLLLAGYDDFNCNVWDALKADRAGVLAGHDNRVSCLGVTDDGMAVATGSWDSFLKIWNGSSGGGGSGGGGSSGVSGWRLFKKIS
>C
NTASIAQARKLVEQLKMEANIDRIKVSKAAADLMAYCEAHAKEDPLLTPVPASENPFRE
>N
QVQLQESGGGLVQPGGSLRLSCAASGFTFSNYKMNWVRQAPGKGLEWVSDISQSGASISYTGSVKGRFTISRDNAKNTLYLQMNSLKPEDTAVYYCARCPAPFTRDCFDVTSTTYAYRGQGTQVTVSS
>R
MTSDFSPEPPMELCYENVNGSCIKSSYAPWPRAILYGVLGLGALLAVFGNLLVIIAILHFKQLHTPTNFLVASLACADFLVGVTVMPFSTVRSVESCWYFGESYCKFHTCFDTSFCFASLFHLCCISIDRYIAVTDPLTYPTKFTVSVSGLCIALSWFFSVTYSFSIFYTGANEEGIEELVVALTCVGGCQAPLNQNWVLLCFLLFFLPTVVMVFLYGRIFLVAKYQARKIEGTANQAQASSESYKERVAKRERKAAKTLGIAMAAFLVSWLPYIIDAVIDAYMNFITPAYVYEILVWCVYYNSAMNPLIYAFFYPWFRKAIKLIVSGKVFRADSSTTNLFSEEAGAG
>S
MLLVNQSHQGFNKEHTSKMVSAIVLYVLLAAAAHSAFAVQLVESGGGLVQPGGSRKLSCSASGFAFSSFGMHWVRQAPEKGLEWVAYISSGSGTIYYADTVKGRFTISRDDPKNTLFLQMTSLRSEDTAMYYCVRSIYYYGSSPFDFWGQGTTLTVSAGGGGSGGGGSGGGGSADIVMTQATSSVPVTPGESVSISCRSSKSLLHSNGNTYLYWFLQRPGQSPQLLIYRMSNLASGVPDRFSGSGSGTAFTLTISRLEAEDVGVYYCMQHLEYPLTFGAGTKLEL
2.2 统计与清洗 CSV 文件
调用 p2_main_gpcr_generate_csv.py,根据 FASTA 文件,在原文件中,去除错误的 Case,保留现有的 Case,即:
python3 gpcr/p2_main_gpcr_generate_csv.py \
-i gpcr/gpcr_data_filter.csv \
-f gpcr/gpcr_complex_v2/fasta_complex_446 \
-o gpcr/gpcr_complex_v2/gpcr_info_446.csv
GPCR 复合物是否包含 抗体(antibody) 链的数据类型:
{'antibody': 340, 'no antibody': 106}
更新的 GPCR 信息文件:
pdb,ab,chains,gpcr,g_len,seqs
7XTC,True,"A,B,G,N,R",R,576,"MGCLGNSKTED..."
2.3 拆分 GPCR 单链的 PDB 与 FASTA
调用 p3_main_gpcr_chain_pdb_exporter.py,根据 GPCR 链拆分 PDB 与 FASTA,即:
python3 gpcr/p3_main_gpcr_chain_pdb_exporter.py \
-i gpcr/gpcr_complex_v2/gpcr_info_446.csv \
-f gpcr/gpcr_complex_v2/fasta_complex_446 \
-p gpcr/gpcr_complex_v2/pdb_complex_446 \
-of gpcr/gpcr_complex_v2/gcpr_chain/fasta \
-op gpcr/gpcr_complex_v2/gcpr_chain/pdb
在拆分 PDB 单链之后,使用格式化 PDB,即保证残基中 CA 只有1个,同时链 ID 转换成 A,有利于后续的结构评估。
输出的 GPCR 链,序列是全长,FASTA 长度大于 PDB 长度,如下:
8IW9_R309.pdb8IW9_R348.fasta
8IW9_R348.fasta 的序列,即:
>R
MTSDFSPEPPMELCYENVNGSCIKSSYAPWPRAILYGVLGLGALLAVFGNLLVIIAILHFKQLHTPTNFLVASLACADFLVGVTVMPFSTVRSVESCWYFGESYCKFHTCFDTSFCFASLFHLCCISIDRYIAVTDPLTYPTKFTVSVSGLCIALSWFFSVTYSFSIFYTGANEEGIEELVVALTCVGGCQAPLNQNWVLLCFLLFFLPTVVMVFLYGRIFLVAKYQARKIEGTANQAQASSESYKERVAKRERKAAKTLGIAMAAFLVSWLPYIIDAVIDAYMNFITPAYVYEILVWCVYYNSAMNPLIYAFFYPWFRKAIKLIVSGKVFRADSSTTNLFSEEAGAG
2.4 GPCR 数据集区分 Monomer 与 Multimer
调用 p4_main_gpcr_monomer_multimer_spliter.py,拆分成 Monomer 与 Multimer 的 FASTA:
python3 gpcr/p4_main_gpcr_monomer_multimer_spliter.py \
-i gpcr/gpcr_complex_v2/fasta_complex_446/ \
-mo gpcr/gpcr_complex_v2/fasta_monomer_36 \
-mu gpcr/gpcr_complex_v2/fasta_multimer_410
确保 Monomer 与 Multimer 的数量之和,等于之前的结构数量,即 36 + 410 = 446,输出如下:
- Monomer 的 FASTA 文件:
8I2H_A682.fasta - Multimer 的 FASTA 文件:
8IW9_A362_B377_C59_N128_R348_S285_1559.fasta
8I2H_A682.fasta 序列如下:
>A
LGSGCHHRICHCSNRVFLCQESKVTEIPSDLPRNAIELRFVLTKLRVIQKGAFSGFGDLEKIEISQNDVLEVIEADVFSNLPKLHEIRIEKANNLLYINPEAFQNLPNLQYLLISNTGIKHLPDVHKIHSLQKVLLDIQDNINIHTIERNSFVGLSFESVILWLNKNGIQEIHNCAFNGTQLDELNLSDNNNLEELPNDVFHGASGPVILDISRTRIHSLPSYGLENLKKLRARSTYNLKKLPTLEKLVALMEASLTYPSHCCAFANWRRQISELHPICNKSILRQEVDYMTQARGQRSSLAEDNESSYSRGFDMTYTEFDYDLCNEVVDVTCSPKPDAFNPCEDIMGYNILRVLIWFISILAITGNIIVLVILTTSQYKLTVPRFLMCNLAFADLCIGIYLLLIASVDIHTKSQYHNYAIDWQTGAGCDAAGFFTVFASELSVYTLTAITLERWHTITHAMQLDCKVQLRHAASVMVMGWIFAFAAALFPIFGISSYMKVSICLPMDIDSPLSQLYVMSLLVLNVLAFVVICGCYIHIYLTVRNPNIVSSSSDTRIAKRMAMLIFTDFLCMAPISFFAISASLKVPLITVSKAKILLVLFHPINSCANPFLYAIFTKNFRRDFFILLSKCGCYEMQAQIYRTETSSTVHNTHPRNGHCSSAPRVTNGSTYILVPLSHLAQN
拆分成 Monomer 与 Multimer 主要原因是蛋白质结构预测算法,区分 Monomer 与 Multimer,使用不同的模型与算法框架,进行预测。
2.5 GPCR 单链区分 Antibody
调用 p5_main_gpcr_antibody_spliter.py,通过标签文件,拆分出 Antibody 与 Non-Antibody 数据集的 PDB 与 FASTA 文件:
python3 gpcr/p5_main_gpcr_antibody_spliter.py \
-i gpcr/gpcr_complex_v2/gpcr_info_446.csv \
-r gpcr/gpcr_complex_v2/gcpr_chain/pdb_446 \
-oa gpcr/gpcr_complex_v2/gcpr_chain/pdb_ab \
-on gpcr/gpcr_complex_v2/gcpr_chain/pdb_nab \
-m pdb
日志:[Info] sample: 446, ab: 340, nab: 106
2.6 从 PDB 中导出已预测残基序列
调用 p6_main_gpcr_short_fasta_exporter.py,从 PDB 中提取 FASTA (短序列),用于后续评估:
python3 gpcr/p6_main_gpcr_short_fasta_exporter.py \
-i gpcr/gpcr_complex_v2/pdb_complex_446 \
-o gpcr/gpcr_complex_v2/fasta_from_pdb_446
PDB 中提取 FASTA 短序列的预测效果,低于真实的长序列,用于后续评估。
输出的 FASTA 序列长度 与 PDB 一致:
- FASTA:
8IW9_A234_B338_C53_R309_S231_N128_1293.fasta - PDB:
8IW9_A234_B338_C53_N128_R309_S231_1293.pdb
3. 搜索 MSA 序列与导出 PDB 结构
搜索 MSA 序列,预测 PDB 结构,导出 PDB 结构,从 Multimer 拆出单链,再根据 FASTA 格式化。
3.1 搜索 MSA
推理 AF2 的 MSA 脚本,需要区分 Monomer 与 Multimer,即:
nohup bash run_alphafold.sh -f gpcr/gpcr_complex_v2/fasta_multimer_410/ -o gpcr/gpcr_complex_v2/fasta_multimer_410_msas -m multimer -h true > nohup.fasta_multimer_410_msas.out &
nohup bash run_alphafold.sh -f gpcr/gpcr_complex_v2/fasta_monomer_36/ -o gpcr/gpcr_complex_v2/fasta_monomer_36_msas -m monomer -h true > nohup.fasta_monomer_36_msas.out &
nohup bash run_alphafold.sh -f gpcr/gpcr_complex_v2/gcpr_chain/fasta_446 -o gpcr/gpcr_complex_v2/gcpr_chain/fasta_446_msas -m monomer -h true > nohup.gcpr_chain_fasta_msas.out &
使用 MSA 服务推理序列,即:
python msa_main.py -m 0 -f gpcr/gpcr_complex_v2/fasta_multimer_410/ -r mydata/test_fasta_multimer_410.json
python msa_main.py -m 1 -r mydata/test_fasta_multimer_410.json
python msa_main.py -m 0 -f gpcr/gpcr_complex_v2/fasta_monomer_36/ -r mydata/test_fasta_monomer_36.json
python msa_main.py -m 1 -r mydata/test_fasta_monomer_36.json
python msa_main.py -m 0 -f gpcr/gpcr_complex_v2/gcpr_chain/fasta_446/ -r mydata/test_fasta_446.json
python msa_main.py -m 1 -r mydata/test_fasta_446.json
以及预测 Monomer 与 Multimer 的结构。
3.1 导出 Monomer 的 PDB 结构
调用 p7_main_xtrimo_monomer_exporter.py,导出预测的 Monomer PDB 结构至单个文件夹,用于后续评估:
python3 gpcr/main_xtrimo_monomer_exporter.py \
-i gpcr_protein_no_antibody/ \
-o mydata/gpcr_eval/baseline_nab_local \
-f gpcr/gpcr_complex/gcpr_chain/fasta_non_ab_95/ \
-r mydata/gpcr_eval/baseline_nab_local_format
3.3 导出 Multimer 的 PDB 结构
调用 p8_main_xtrimo_multimer_exporter.py,导出预测的 Multimer PDB 结构至单个文件夹,用于后续评估:
python3 gpcr/main_xtrimo_multimer_exporter.py \
-i mydata/outputs_infer/gpcr_fasta_multimer_392_outputs/ \
-o mydata/gpcr_eval/gpcr_fasta_multimer_392 \
-f gpcr/gpcr_complex/fasta_multimer_392/ \
-r mydata/gpcr_eval/gpcr_fasta_multimer_392_format
再调用 p9_main_multimer_to_chain_exporter.py,从 Multimer 中 导出 GPCR 单链结构,即:
python3 gpcr/p9_main_multimer_to_chain_exporter.py \
-i mydata/gpcr_eval/gpcr_fasta_multimer_392_format \
-o mydata/gpcr_eval/gpcr_chain_multimer_v1 \
-c gpcr/gpcr_complex_v2/gpcr_info_446.csv
Monomer 的单链与 Multimer 导出的单链,都需要根据 PDB 序列去除冗余残基,只保留与 PDB 一致的残基。
调用 p10_main_pdb_residues_format.py,预测 PDB 结构的序列,匹配目标 PDB 结构的序列,标准化格式,用于之后的单链评估,即:
python3 gpcr/p10_main_pdb_residues_format.py \
-i mydata/gpcr_eval/gpcr_chain_redundancy_multimer \
-o mydata/gpcr_eval/gpcr_chain_redundancy_multimer_format \
-f gpcr/gpcr_complex_v2/fasta_from_pdb_446
4. 评估模型性能
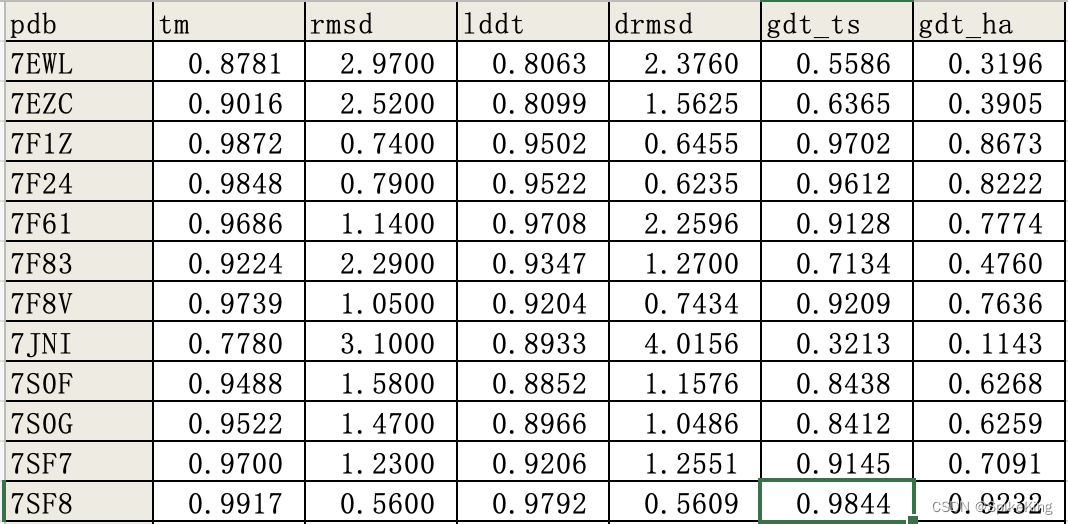
调用 p11_main_gpcr_evaluator.py 评估预测结果,支持 ["pdb", "tm", "rmsd", "lddt", "drmsd", "gdt_ts", "gdt_ha"]
- 输入文件夹
-m - 实验名称
-n - 真实结构文件夹
-t - 输出 xls 文件
-o
即:
python3 gpcr/p11_main_gpcr_evaluator.py \
-m mydata/gpcr_eval/gpcr_chain_redundancy_multimer_format \
-n nab_multimer_redundancy \
-t mydata/gpcr_targets/pdb_non_ab_95_format \
-o mydata/gpcr_res/
输出效果:
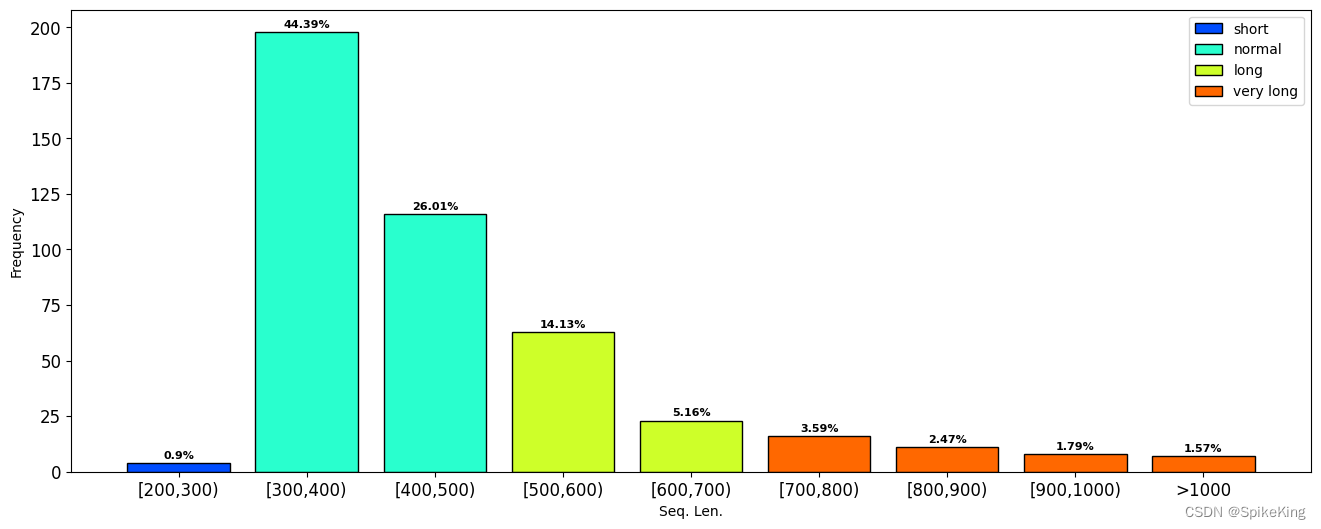
调用 p12_main_gpcr_info_analysis.py 统计信息,支持展示序列长度:
python3 gpcr/p12_main_gpcr_info_analysis.py \
-i gpcr/gpcr_complex_v2/gpcr_info_446.csv \
-o mydata/gpcr/gpcr_images \
-k g_len
日志:
[Info] seq len range: 283 ~ 1543
[Info] len > 20: 446, len < 20: 0
[Info] value_counts: 300: 198 (44.3946%), 400: 116 (26.009%), 500: 63 (14.1256%), 600: 23 (5.157%), 700: 16 (3.5874%), 800: 11 (2.4664%), 900: 8 (1.7937%), 1000: 7 (1.5695%), 200: 4 (0.8969%), sum: 446
效果: