GCE的安装和使用
- GCE的安装使用
- 1. GCE的安装
- 2. GCE的使用
- 补充:一个简单的R脚本——kmerpdf.R,用于绘制kmer的种类和数量分布图
GCE的安装使用
一个基因组评估软件。其他同类型软件Genomescope
1. GCE的安装
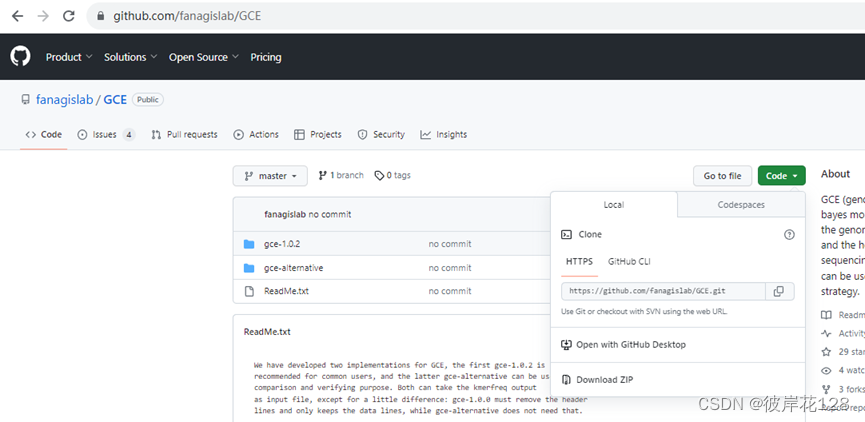
Github官网:https://github.com/fanagislab/GCE
点击Code,点击Download ZIP,下载文件GCE-master.zip
 cd /home/zhaohuiyao/Biosoft
cd /home/zhaohuiyao/Biosoft
#上传文件GCE-master.zip
unzip GCE-master.zip
cd GCE-master/
make
#安装成功
#可执行文件位置:/home/zhaohuiyao/Biosoft/GCE-master/gce-1.0.2/gce
2. GCE的使用
准备好过滤后的二代cleandata
cd /home/zhaohuiyao/Genome_survey/03kmer
#创建cleandata.txt文件

#初步设定kmer=17mer(常用)
#kmer频率统计
/home/zhaohuiyao/Biosoft/GCE-master/gce-1.0.2/kmerfreq -k 17 -t 18 -p 17mer_shuxi cleandata.txt
awk ‘{if($0~/^#/ || $0 == “”); else{ print $0}}’ 17mer_shuxi.kmer.freq.stat | cut -f 1,2 > 17mer_shuxi.kmer.freq.stat.2colum
#在文件17mer_shuxi.kmer.freq.stat中查看kmer的种类数目——作为GCE参数-g的值40451775089
grep “#Kmer indivdual number” 17mer_shuxi.kmer.freq.stat
#GCE评估(第一次)
/home/zhaohuiyao/Biosoft/GCE-master/gce-1.0.2/gce -f 17mer_shuxi.kmer.freq.stat.2colum -g 40451775089 >17mer_shuxi.table 2> 17mer_shuxi.log
#GCE评估(第二次)
/home/zhaohuiyao/Biosoft/GCE-master/gce-1.0.2/gce -f 17mer_shuxi.kmer.freq.stat.2colum -g 40451775089 -H 1 -c 64 >17mer_shuxi_2.table 2> 17mer_shuxi_2.log
#在文件17mer_shuxi_2.log文件中查看以下信息
grep -A 1 “raw_peak” 17mer_shuxi_2.log

#依据genome_size、a[1/2]、a[1]、b[1/2]、b[1]值
#计算重复序列占比R=1-b[1/2]-b[1]=1-0.101873-0.279256=61.89%,杂合度H=[a[1/2]/(2-a[1/2])]/kmer_value=[0.270554/(2-0.270554)]/17=0.92%
#若计算的杂合度H<0.5%,则表示该物种是纯合物种,那么重复序列占比R需要重新计算,使用文件17mer_shuxi.log中的信息。R=1-b[1]=1-0.431343=56.87%

#因为杂合度H=0.92%≥0.5%,所以测序物种为杂合种。不需要重新计算重复序列占比R。
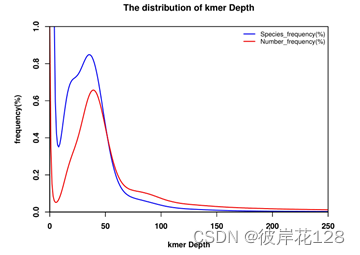
补充:一个简单的R脚本——kmerpdf.R,用于绘制kmer的种类和数量分布图
Rscript kmerpdf.R 17mer_shuxi.kmer.freq.stat.2colum 17mer_shuxi.kmer.freq.pdf 40451775089
args <- commandArgs(T)
df <- read.table(args[1])
colnames(df) <- c("kmer Depth","Species")
df$'Number'<- df$'kmer Depth'*df$'Species'
df$'Species_frequency(%)'=df$'Species'*100/sum(df$'Species')
df$'Number_frequency(%)'=df$'Number'*100/as.numeric(args[3])
Spefreq_peak=round(max(df$'Species_frequency(%)'[10:nrow(df)])+0.2,1)
Numfreq_peak=round(max(df$'Number_frequency(%)'[10:nrow(df)])+0.2,1)
pdf(args[2],height=8,width=10)
par(pin=c(6,4))
plot(x=df$'kmer Depth',y=df$'Species_frequency(%)',type="l",xlim=c(0,250),ylim=c(0,Spefreq_peak),col="blue2",
lwd=2,xlab="",ylab="",xaxs = 'i',yaxs = 'i',main="The distribution of kmer Depth")
mtext("Species_frequency(%)",side=2,line = 3,font=2);mtext("kmer Depth",side=1,line = 3,font=2)
axis(side=1,line=0,font=2);axis(side=2,line=0,font=2)
par(new=TRUE)
plot(x=df$'kmer Depth',y=df$'Number_frequency(%)',type="l",xlim=c(1,250),ylim=c(0,Numfreq_peak),col="red2",
lwd=2,xlab="",ylab="",xaxs = 'i',yaxs = 'i',xaxt="n",yaxt="n")
legend("topright",legend=c("Species_frequency(%)","Number_frequency(%)"),
col=c("blue2","red2"),lty=1,lwd=2,cex=0.8,bty="n")
dev.off()
#结果文件17mer_shuxi.kmer.freq.pdf