cs224w_colab2.py这个图属性预测到底咋预测的
dataset.meta_info.T
Out[2]:
num tasks 1
eval metric rocauc
download_name hiv
version 1
url http://snap.stanford.edu/ogb/data/graphproppre...
add_inverse_edge True
data type mol
has_node_attr True
has_edge_attr True
task type binary classification
num classes 2
split scaffold
additional node files None
additional edge files None
binary False
Name: ogbg-molhiv, dtype: object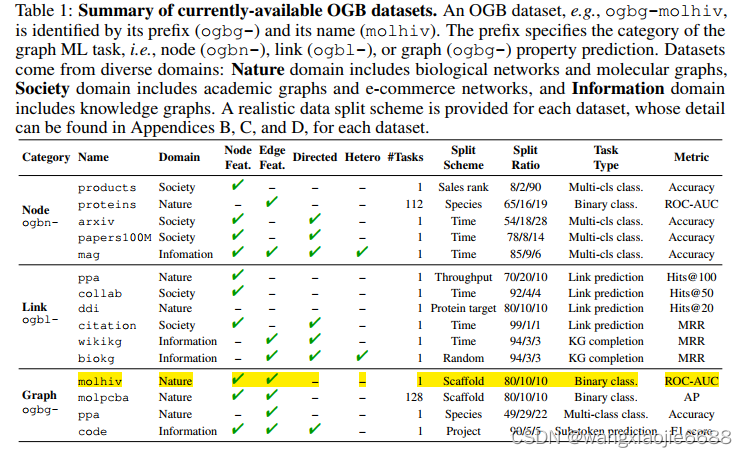
 参照上面 这里的num tasks 仅适用于图属性预测? num tasks = 1
参照上面 这里的num tasks 仅适用于图属性预测? num tasks = 1
model = GCN_Graph(args['hidden_dim'],
dataset.num_tasks, args['num_layers'],
args['dropout']).to(device)
train_loader.dataset.data.edge_index.shape
Out[10]: torch.Size([2, 2259376])
train_loader.dataset.data.edge_attr.shape
Out[12]: torch.Size([2259376, 3])
type(train_loader.dataset.data.node_stores)
Out[26]: list
train_loader.dataset.data.node_stores[0]['y'].shape
Out[46]: torch.Size([41127, 1])
train_loader.dataset.data.node_stores[0]['y'].sum()
Out[47]: tensor(1443) y 中的数值求和值
torch.unique(train_loader.dataset.data.node_stores[0]['y'],return_counts=True)
Out[58]: (tensor([0, 1]), tensor([39684, 1443])) 仅0,1两类
self.node_encoder.atom_embedding_list
Out[62]:
ModuleList(
(0): Embedding(119, 256)
(1): Embedding(5, 256)
(2): Embedding(12, 256)
(3): Embedding(12, 256)
(4): Embedding(10, 256)
(5): Embedding(6, 256)
(6): Embedding(6, 256)
(7): Embedding(2, 256)
(8): Embedding(2, 256)
)
list(enumerate(data_loader))
Out[82]:
[(0,
DataBatch(edge_index=[2, 1734], edge_attr=[1734, 3], x=[807, 9], y=[32, 1], num_nodes=807, batch=[807], ptr=[33])),
若干组 很多 x, edge_index, batch = batched_data.x, batched_data.edge_index, batched_data.batch
embed = self.node_encoder(x) #使用编码器 将原先9维的编码为256维 self.node_encoder = AtomEncoder(hidden_dim)
out = self.gnn_node(embed, edge_index) #使用gcn得到节点嵌入 embed=X edge_index 连边/节点对
out = self.pool(out, batch)
batch.unique(return_counts = True)
Out[94]:
(tensor([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31],
device='cuda:0'), 这里说明有31个待训练子图(化学分子) 下图api说明了聚合过程
tensor([30, 18, 21, 26, 12, 20, 17, 18, 36, 11, 31, 22, 21, 26, 22, 21, 21, 63,
15, 18, 18, 29, 18, 40, 41, 19, 19, 30, 12, 21, 19, 23], 每个分子中所含有的节点(原子)数量
device='cuda:0'))
batch.shape
Out[95]: torch.Size([758])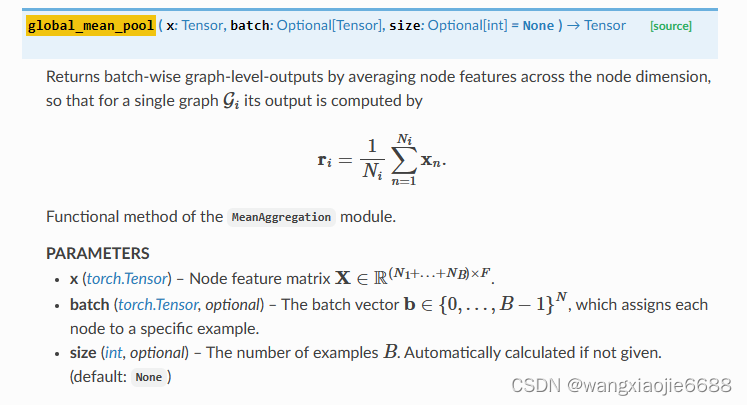
def global_mean_pool(x: Tensor, batch: Optional[Tensor],
size: Optional[int] = None) -> Tensor:
dim = -1 if x.dim() == 1 else -2 #这里的x.dim() = 2
# dim() → int Returns the number of dimensions of self tensor.
if batch is None:
return x.mean(dim=dim, keepdim=x.dim() <= 2) #keepdim=x.dim() <= 2 ??啥玩意<=
size = int(batch.max().item() + 1) if size is None else size
return scatter(x, batch, dim=dim, dim_size=size, reduce='mean')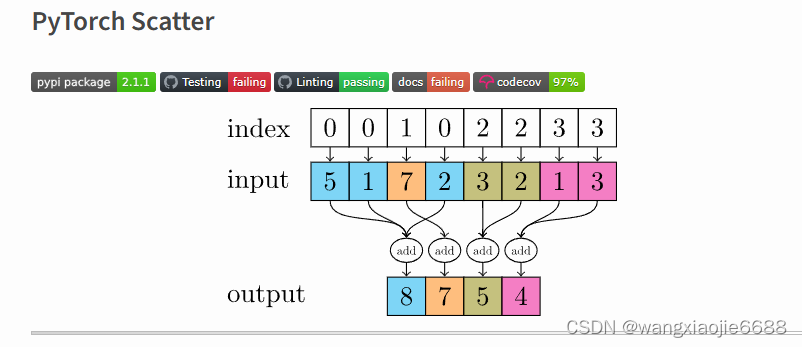
This package consists of a small extension library of highly optimized sparse update (scatter and segment) operations for the use in PyTorch, which are missing in the main package. Scatter and segment operations can be roughly described as reduce operations based on a given "group-index" tensor. Segment operations require the "group-index" tensor to be sorted, whereas scatter operations are not subject to these requirements.该包由一个小型扩展库组成,该库包含用于PyTorch的高度优化的稀疏更新(分散和分段)操作,这些操作在主包中丢失。散射和分段运算可以粗略地描述为基于给定“群索引”张量的归约运算。分段运算需要对“组索引”张量进行排序,而分散运算则不受这些要求的约束。
由此(scatter)由多个节点的嵌入值最终得到这部分节点所在的子图嵌入(化学分子)。
def forward(self, batched_data):
# TODO: Implement a function that takes as input a
# mini-batch of graphs (torch_geometric.data.Batch) and
# returns the predicted graph property for each graph.
#
# NOTE: Since we are predicting graph level properties,
# your output will be a tensor with dimension equaling
# the number of graphs in the mini-batch
x, edge_index, batch = batched_data.x, batched_data.edge_index, batched_data.batch
embed = self.node_encoder(x) #使用编码器 将原先9维的编码为256维 self.node_encoder = AtomEncoder(hidden_dim)
out = self.gnn_node(embed, edge_index) #使用gcn得到节点嵌入 embed=X edge_index 连边/节点对
out = self.pool(out, batch)
out = self.linear(out)
############# Your code here ############
## Note:
## 1. Construct node embeddings using existing GCN model
## 2. Use the global pooling layer to aggregate features for each individual graph
## For more information please refer to the documentation:
## https://pytorch-geometric.readthedocs.io/en/latest/modules/nn.html#global-pooling-layers
## 3. Use a linear layer to predict each graph's property
## (~3 lines of code)
#########################################
return out
out.shape
Out[122]: torch.Size([32, 1])
out
Out[121]:
tensor([[-0.4690],
[-1.0285],
[-0.4614],
最后经过线性层 返回得到所属类别概率 运行到如下部分结束反向传播forward() (op = model(batch)# 先进入model函数 然后运行 反向传播)
def train(model, device, data_loader, optimizer, loss_fn):
# TODO: Implement a function that trains your model by
# using the given optimizer and loss_fn.
model.train() #Sets the module in training mode. data_loader.dataset.data Data(num_nodes=1049163, edge_index=[2, 2259376], edge_attr=[2259376, 3], x=[1049163, 9], y=[41127, 1])
loss = 0
for step, batch in enumerate(tqdm(data_loader, desc="Iteration")): #,total= data_loader.batch_sampler
# for step, batch in tqdm(enumerate(data_loader), desc="Iteration"): #,total= data_loader.batch_sampler
batch = batch.to(device)
if batch.x.shape[0] == 1 or batch.batch[-1] == 0:
pass
else:
## ignore nan targets (unlabeled) when computing training loss.
is_labeled = batch.y == batch.y # 0/1转化为Ture/False
############# Your code here ############
## Note:
## 1. Zero grad the optimizer
## 2. Feed the data into the model
## 3. Use `is_labeled` mask to filter output and labels
## 4. You may need to change the type of label to torch.float32
## 5. Feed the output and label to the loss_fn
## (~3 lines of code)
optimizer.zero_grad()
# print('optimizer.zero_grad()')
op = model(batch)# 先进入model函数 然后运行 反向传播
。。。。。。。。。。。。。。。后面计算损失 更新梯度等等存在错误等欢迎指正! 附件为整个作业的.py文件