很久以前,我们发布过一个单细胞多组差异基因可视化的方法。跟着Cell学单细胞转录组分析(八):单细胞转录组差异基因分析及多组结果可视化。主要复现参考的是这篇发表在《Cell》上的文章。可以将多个组的差异结果展示出来。
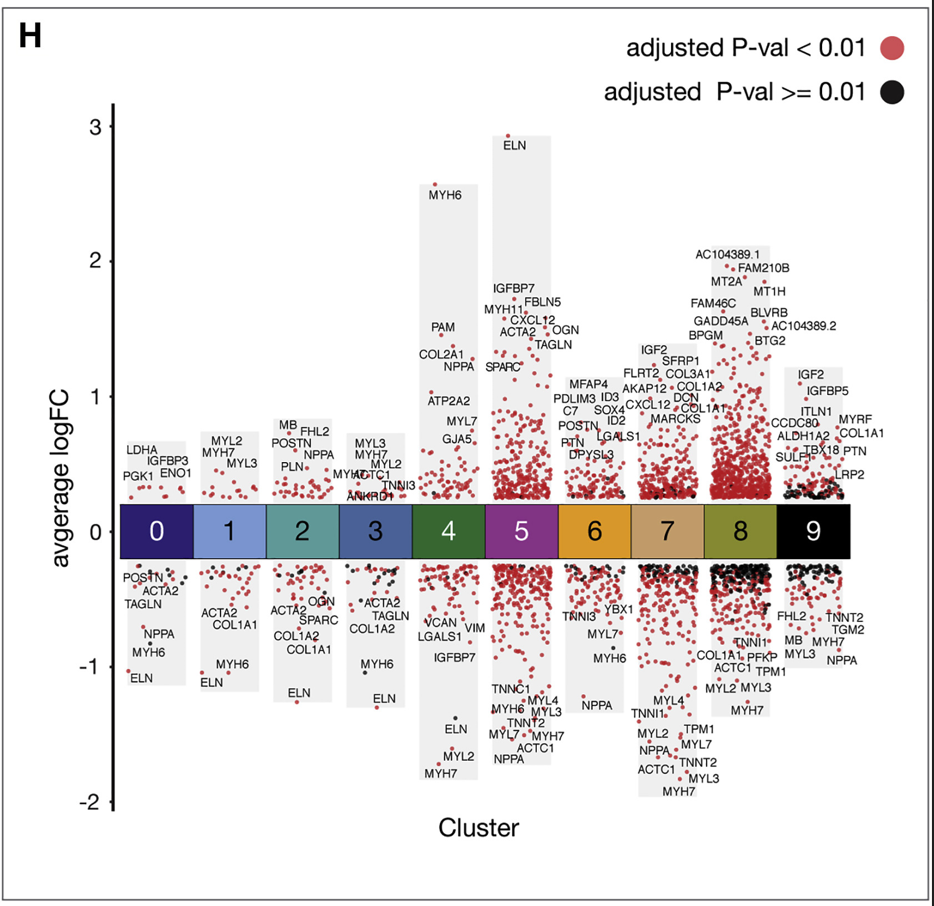
(reference:A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart)
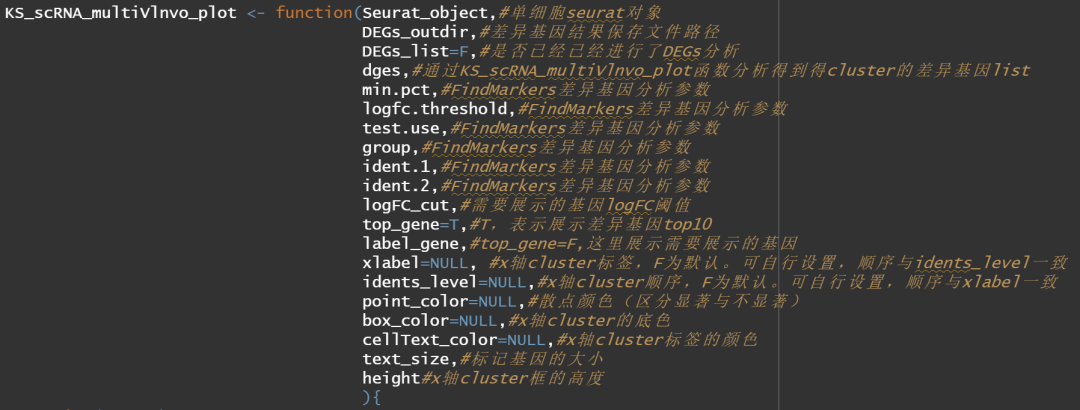
这里我们对之前的帖子进行更新,主要是解决之前的一些问题,让作图更加清晰正确。此外,我们直接将差异基因的分析和可视化包装成一个通用的函数,函数有一定的可调节性。
函数解释如下:
函数的使用视频见B站:
https://www.bilibili.com/video/BV19w411D7qZ/?spm_id_from=333.999.0.0
接下来我们测试一下:这里差异基因的分析使用的是Seurat的Findmarkers函数,所以一些参数和Findmarkers是一样的,自行调节。最好可以将logfc.threshold和min.pct设置为0,这样就可以获得所有的基因,这个结果我们函数是直接保存在相关路径中的,那么这么做有什么用呢?这个结果可以进行富集分析,更重要的是可以进行GSEA分析(复现Nature图表:GSEA分析及可视化包装函数、(视频教程)GSEA分析可视化函数/棒棒糖图展示富集结果)。
KS_scRNA_multiVlnvo_plot(Seurat_object = uterus,DEGs_outdir = "./",DEGs_list=F,min.pct = 0.3,logfc.threshold = 0.3,test.use = "wilcox",group = "orig.ident",ident.1 = "EEC",ident.2 = "HC",logFC_cut = 0.3,top_gene=T,text_size = 3,height = 0.3)
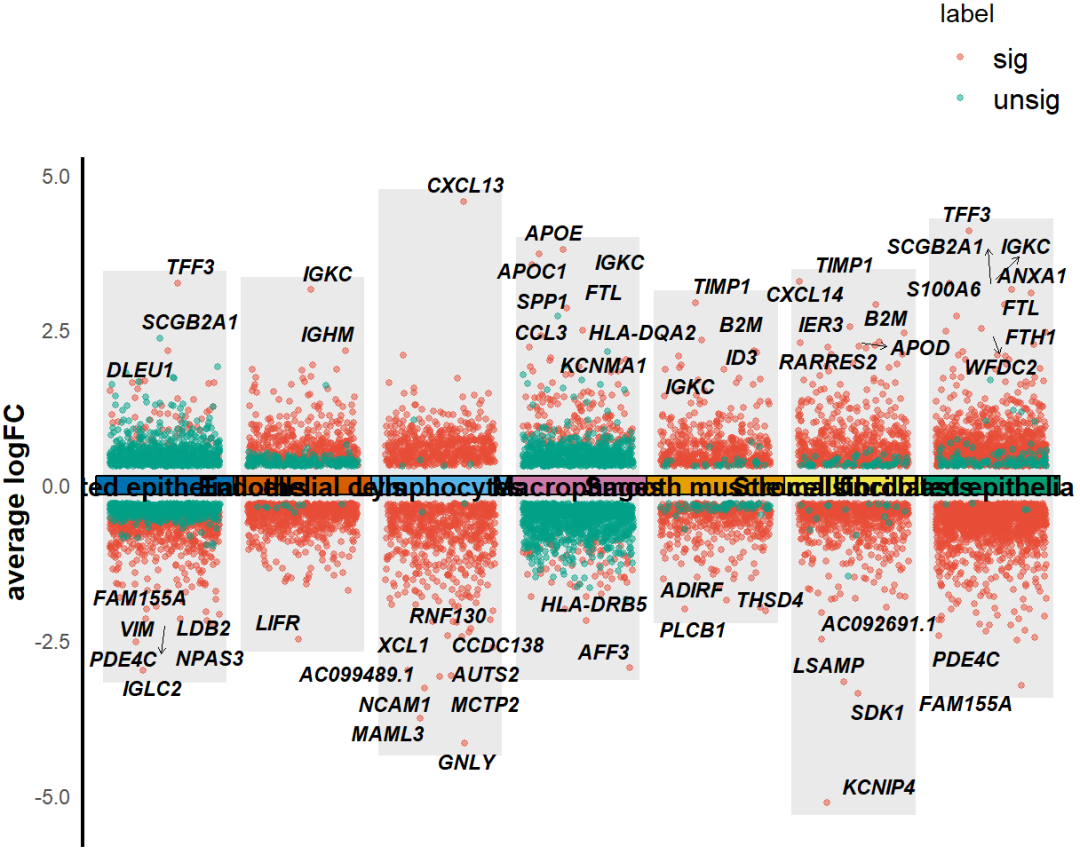
很显然,由于不知道差异结果如何,所以上面的图参数没有调整,不是很好,我们需要进行细节调整。可是如果是这样的话,难道又要跑一遍差异基因分析吗?显然是很麻烦的,我们可考虑到这个问题。还记得上一步我们差异基因结果已经保存了吗,接下来只需要作图的话dges参数设置为刚才保存的差异结果,DEGs_list设置为T。
KS_scRNA_multiVlnvo_plot(Seurat_object = uterus,DEGs_list=T,dges = sce_DEGs,logFC_cut = 0.3,top_gene=T,text_size = 3,height = 0.8)
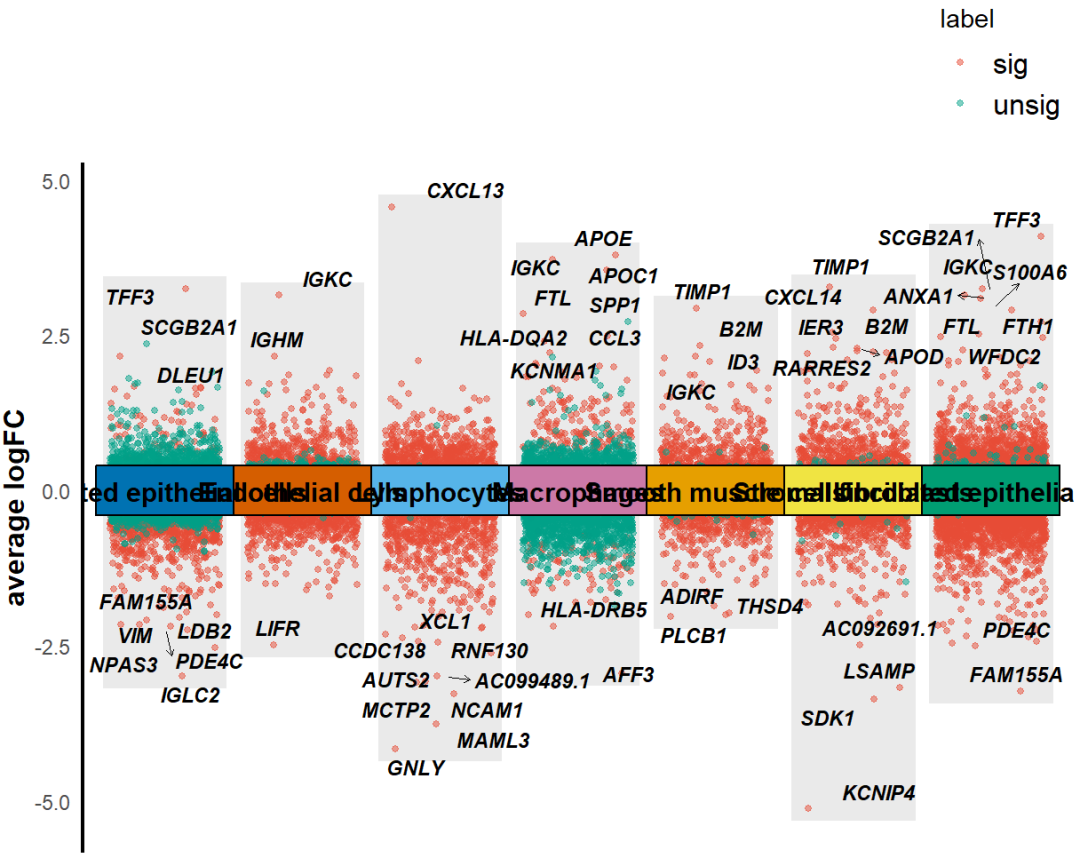
然后,我们还可以调整x轴cluster的顺序,调整label。
KS_scRNA_multiVlnvo_plot(Seurat_object = uterus,DEGs_list=T,dges = sce_DEGs,logFC_cut = 0.3,top_gene=T,text_size = 3,xlabel = c("SMC","Ly","UEC","SF","CEP","EC","MAC"),idents_level = c("Smooth muscle cells","Lymphocytes","Unciliated epithelial cells","Stromal fibroblasts","Ciliated epithelial cells","Endothelial cells","Macrophages"),height = 0.8)

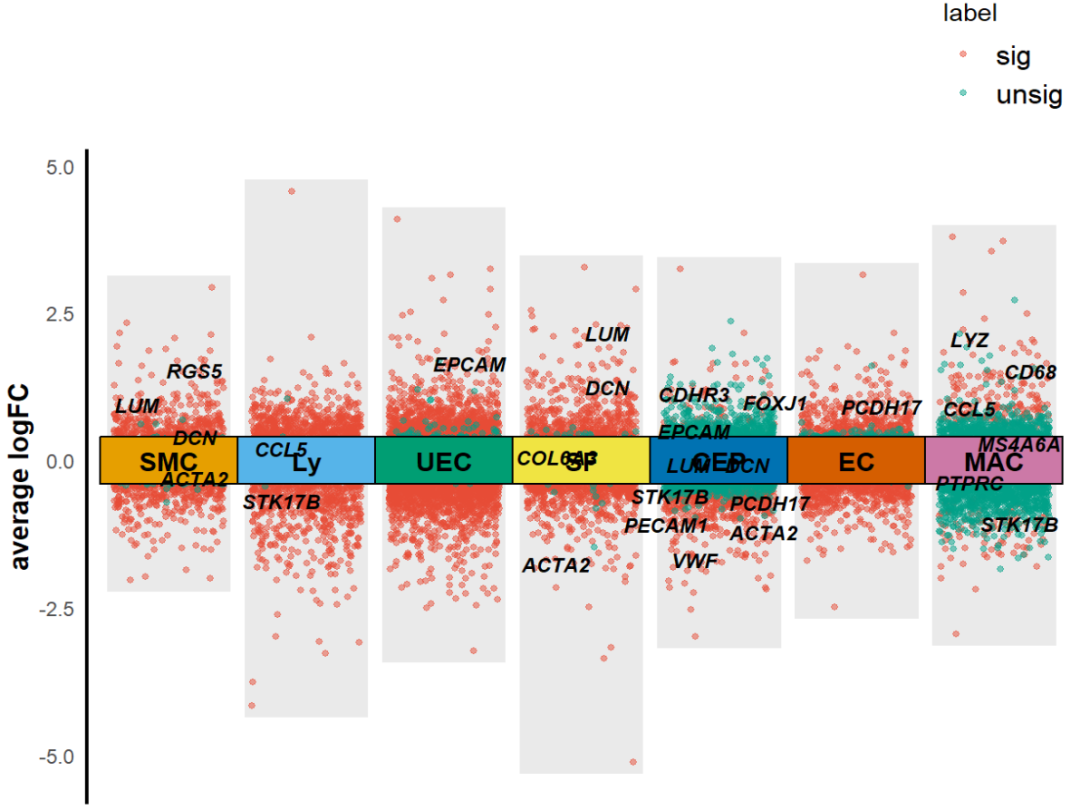
最后,标记的基因也可以自选。
KS_scRNA_multiVlnvo_plot(Seurat_object = uterus,DEGs_list=T,dges = sce_DEGs,logFC_cut = 0.3,text_size = 3,xlabel = c("SMC","Ly","UEC","SF","CEP","EC","MAC"),idents_level = c("Smooth muscle cells","Lymphocytes","Unciliated epithelial cells","Stromal fibroblasts","Ciliated epithelial cells","Endothelial cells","Macrophages"),height = 0.8,top_gene = F,label_gene = c("ACTA2", "RGS5","MS4A6A", "CD68","LYZ","CCL5", "STK17B","PTPRC","DCN", "COL6A3","LUM","PECAM1","PCDH17", "VWF","EPCAM","CDH1","FOXJ1","CDHR3","DYDC2"))
好了,这就是这个函数的所有内容了,如果觉得分享有用,点个赞再走呗!






![[ES]安装es、kibana、ik分词器](https://img-blog.csdnimg.cn/367d53f3587a4ca38300808d2ed38685.png)