甲基化系列分析教程
桓峰基因公众号推出甲基化系列分析教程,整理如下:
甲基化系列 1. 甲基化之前世今生(Methylation)
甲基化系列 2. 甲基化芯片数据介绍与下载(GEO)
甲基化系列 3. 甲基化芯片数据分析完整版(ChAMP)
甲基化系列的教程有很久没有更新了,有这方研究方向的高手,有偿约稿,辛苦费大大地有,速速联系我吧!
简介
DNA 甲基化特征通常是基于多元的方法,需要数以百计的预测网站。在这里,我们提出了一个计算框架 CimpleG 用于检测 CpG 甲基化特征,用于细胞类型分类功能和反卷积。CimpleG 既具有时间效率,又具有执行能力以及表现最好的血细胞和其他细胞类型分类方法而它的预测是基于每种细胞类型的单个 DNA 甲基化位点。总之,CimpleG 为描述提供了一个完整的计算框架 DNA 特征和细胞反褶积。

(A) 对用于特性选择和下游应用程序的 CimpleG 和统计的概述。体细胞(B)或白细胞(C)的DNAm数据集的主成分分析。训练样本(≥10个阳性样本)和测试数据的粗体突出显示的细胞类型被用作目标类。测试数据中不存在的细胞类型仅用作阴性示例(非靶细胞)。

软件包安装
# Install directly from github:
devtools::install_github("costalab/CimpleG")
# Alternatively, downloading from our release page and installing it from a local source:
# - ie navigating through your system
install.packages(file.choose(), repos = NULL, type = "source")
# - ie given a path to a local source
install.packages("~/Downloads/CimpleG_0.0.5.XXXX.tar.gz", repos = NULL, type = "source")
# or
devtools::install_local("~/Downloads/CimpleG_0.0.5.XXXX.tar.gz")数据读取
library("CimpleG")
data(train_data)
train_data[1:5,1:5]
## cg10456121 cg01954746 cg07580588 cg11857210 cg02674789
## GSM2759107 0.5357843 0.9270407 0.9571794 0.9016588 0.9556690
## GSM2759108 0.5516279 0.9351165 0.9500683 0.8665362 0.9421682
## GSM2759109 0.5154959 0.9054998 0.9538037 0.9327675 0.8824507
## GSM2759110 0.5887189 0.9108159 0.9551378 0.9156878 0.9249404
## GSM2759111 0.4917143 0.9397178 0.9519388 0.8655511 0.8840323
data(train_targets)
head(train_targets)
## gsm cell_type adipocytes astrocytes blood_cells
## 1 GSM2759107 endothelial_cells 0 0 0
## 2 GSM2759108 endothelial_cells 0 0 0
## 3 GSM2759109 endothelial_cells 0 0 0
## 4 GSM2759110 endothelial_cells 0 0 0
## 5 GSM2759111 endothelial_cells 0 0 0
## 6 GSM2759112 endothelial_cells 0 0 0
## endothelial_cells epidermal_cells epithelial_cells fibroblasts glia
## 1 1 0 0 0 0
## 2 1 0 0 0 0
## 3 1 0 0 0 0
## 4 1 0 0 0 0
## 5 1 0 0 0 0
## 6 1 0 0 0 0
## hepatocytes ips_cells msc muscle_cells neurons muscle_sc group_data
## 1 0 0 0 0 0 0 train
## 2 0 0 0 0 0 0 train
## 3 0 0 0 0 0 0 train
## 4 0 0 0 0 0 0 train
## 5 0 0 0 0 0 0 train
## 6 0 0 0 0 0 0 train
## description
## 1 ENDOTHELIAL.CELLS
## 2 ENDOTHELIAL.CELLS
## 3 ENDOTHELIAL.CELLS
## 4 ENDOTHELIAL.CELLS
## 5 ENDOTHELIAL.CELLS
## 6 ENDOTHELIAL.CELLS
colnames(train_targets)
## [1] "gsm" "cell_type" "adipocytes"
## [4] "astrocytes" "blood_cells" "endothelial_cells"
## [7] "epidermal_cells" "epithelial_cells" "fibroblasts"
## [10] "glia" "hepatocytes" "ips_cells"
## [13] "msc" "muscle_cells" "neurons"
## [16] "muscle_sc" "group_data" "description"
data(test_data)
test_data[1:5,1:5]
## cg10456121 cg01954746 cg07580588 cg11857210 cg02674789
## GSM1289142 0.06305196 0.8088967 0.7291303 0.8387920 0.7728405
## GSM1289143 0.09940250 0.8411065 0.7906977 0.8273305 0.7540370
## GSM1289146 0.10658357 0.8103309 0.7638934 0.8142458 0.7384098
## GSM1289144 0.24916585 0.8149448 0.8164471 0.8223350 0.7968497
## GSM1289145 0.11871330 0.8754153 0.8049503 0.7617512 0.7903106
data(test_targets)
# check the train_targets table to see
# what other columns can be used as targets
# colnames(train_targets)实例操作
CimpleG试图找到对给定的训练数据集的细胞类型进行最佳分类的CpG, 还能够在几个简单的步骤中执行细胞型反褶积,可以使用beta值或M值。这里我们展示了生成signature就非常容易了。
运行 CimpleG
运行CimpleG非常简单。您只需要使用一些参数运行CimpleG函数。
# mini example with just 4 target signatures
set.seed(42)
cimpleg_result <- CimpleG(
train_data = train_data,
train_targets = train_targets,
test_data = test_data,
test_targets = test_targets,
method = "CimpleG",
target_columns = c(
"neurons",
"glia",
"blood_cells",
"fibroblasts"
)
)
## Training for target 'neurons' with 'CimpleG' has finished.: 1.63 sec elapsed
## Training for target 'glia' with 'CimpleG' has finished.: 0.5 sec elapsed
## Training for target 'blood_cells' with 'CimpleG' has finished.: 0.37 sec elapsed
## Training for target 'fibroblasts' with 'CimpleG' has finished.: 0.36 sec elapsed
cimpleg_result$results
## $neurons
## $neurons$train_res
## $neurons$train_res$fold_id
## # A tibble: 4,090 × 3
## Row Data Fold
## <int> <chr> <chr>
## 1 1 Analysis Fold02
## 2 1 Analysis Fold03
## 3 1 Analysis Fold04
## 4 1 Analysis Fold05
## 5 1 Analysis Fold06
## 6 1 Analysis Fold07
## 7 1 Analysis Fold08
## 8 1 Analysis Fold09
## 9 1 Analysis Fold10
## 10 2 Analysis Fold01
## # ℹ 4,080 more rows
##
## $neurons$train_res$train_summary
## id stat_origin mean_aupr mean_var_a fold_presence
## 1: cg02124957 train_aupr 1.0000000 0.03121190 10
## 2: cg02124957 validation_aupr 1.0000000 0.03121190 10
## 3: cg13700051 train_aupr 1.0000000 0.03278101 10
## 4: cg13700051 validation_aupr 1.0000000 0.03278101 10
## 5: cg14356362 train_aupr 0.9787985 0.02450627 10
## 6: cg14356362 validation_aupr 0.9791667 0.02450627 10
## 7: cg21637776 train_aupr 0.9663700 0.04718167 8
## 8: cg21637776 validation_aupr 1.0000000 0.04718167 8
## 9: cg24548498 train_aupr 1.0000000 0.02497980 10
## 10: cg24548498 validation_aupr 1.0000000 0.02497980 10
##
## $neurons$train_res$dt_dmsv
## id diff_means sum_variance pred_type
## 1: cg02124957 -0.5272931 0.008677887 FALSE
## 2: cg13700051 -0.5790852 0.010987231 FALSE
## 3: cg14356362 -0.6419124 0.010098182 FALSE
## 4: cg21637776 -0.3788893 0.006784796 FALSE
## 5: cg24548498 0.5781948 0.008344877 TRUE
##
## $neurons$train_res$train_results
## id fold_presence diff_means sum_variance pred_type mean_var_a
## 1: cg24548498 10 0.5781948 0.008344877 TRUE 0.02497980
## 2: cg14356362 10 -0.6419124 0.010098182 FALSE 0.02450627
## 3: cg02124957 10 -0.5272931 0.008677887 FALSE 0.03121190
## 4: cg13700051 10 -0.5790852 0.010987231 FALSE 0.03278101
## 5: cg21637776 8 -0.3788893 0.006784796 FALSE 0.04718167
## train_aupr validation_aupr cpg_score train_rank
## 1: 1.0000000 1.0000000 0.001998384 1
## 2: 0.9787985 0.9791667 0.002380850 2
## 3: 1.0000000 1.0000000 0.002496952 3
## 4: 1.0000000 1.0000000 0.002622481 4
## 5: 0.9663700 1.0000000 0.005138542 5
##
##
## $neurons$test_perf
## id fold_presence diff_means sum_variance pred_type mean_var_a
## 1: cg24548498 10 0.5781948 0.008344877 TRUE 0.02497980
## 2: cg14356362 10 -0.6419124 0.010098182 FALSE 0.02450627
## 3: cg02124957 10 -0.5272931 0.008677887 FALSE 0.03121190
## 4: cg13700051 10 -0.5790852 0.010987231 FALSE 0.03278101
## 5: cg21637776 8 -0.3788893 0.006784796 FALSE 0.04718167
## train_mean_aupr validation_mean_aupr cpg_score train_rank test_aupr
## 1: 1.0000000 1.0000000 0.001998384 1 0.9215653
## 2: 0.9787985 0.9791667 0.002380850 2 1.0000000
## 3: 1.0000000 1.0000000 0.002496952 3 0.8796205
## 4: 1.0000000 1.0000000 0.002622481 4 0.4853427
## 5: 0.9663700 1.0000000 0.005138542 5 0.6187609
##
## $neurons$elapsed_time
## Time difference of 1.687599 secs
##
##
## $glia
## $glia$train_res
## $glia$train_res$fold_id
## # A tibble: 4,090 × 3
## Row Data Fold
## <int> <chr> <chr>
## 1 1 Analysis Fold01
## 2 1 Analysis Fold02
## 3 1 Analysis Fold03
## 4 1 Analysis Fold05
## 5 1 Analysis Fold06
## 6 1 Analysis Fold07
## 7 1 Analysis Fold08
## 8 1 Analysis Fold09
## 9 1 Analysis Fold10
## 10 2 Analysis Fold01
## # ℹ 4,080 more rows
##
## $glia$train_res$train_summary
## id stat_origin mean_aupr mean_var_a fold_presence
## 1: cg02011981 train_aupr 1.0000000 0.03369357 10
## 2: cg02011981 validation_aupr 1.0000000 0.03369357 10
## 3: cg07644184 train_aupr 0.5982708 0.03479852 10
## 4: cg07644184 validation_aupr 0.7750000 0.03479852 10
## 5: cg11150667 train_aupr 0.7621032 0.04904385 10
## 6: cg11150667 validation_aupr 0.9250000 0.04904385 10
## 7: cg14501977 train_aupr 1.0000000 0.01745930 10
## 8: cg14501977 validation_aupr 1.0000000 0.01745930 10
## 9: cg25737283 train_aupr 0.9556481 0.04227059 10
## 10: cg25737283 validation_aupr 1.0000000 0.04227059 10
##
## $glia$train_res$dt_dmsv
## id diff_means sum_variance pred_type
## 1: cg02011981 -0.5970260 0.012028332 FALSE
## 2: cg07644184 -0.4876997 0.008267879 FALSE
## 3: cg11150667 -0.2643837 0.003424957 FALSE
## 4: cg14501977 -0.6653107 0.007728417 FALSE
## 5: cg25737283 -0.4803594 0.009757117 FALSE
##
## $glia$train_res$train_results
## id fold_presence diff_means sum_variance pred_type mean_var_a
## 1: cg14501977 10 -0.6653107 0.007728417 FALSE 0.01745930
## 2: cg02011981 10 -0.5970260 0.012028332 FALSE 0.03369357
## 3: cg25737283 10 -0.4803594 0.009757117 FALSE 0.04227059
## 4: cg11150667 10 -0.2643837 0.003424957 FALSE 0.04904385
## 5: cg07644184 10 -0.4876997 0.008267879 FALSE 0.03479852
## train_aupr validation_aupr cpg_score train_rank
## 1: 1.0000000 1.000 0.001396744 1
## 2: 1.0000000 1.000 0.002695486 2
## 3: 0.9556481 1.000 0.003825165 3
## 4: 0.7621032 0.925 0.007052477 4
## 5: 0.5982708 0.775 0.009051174 5
##
##
## $glia$test_perf
## id fold_presence diff_means sum_variance pred_type mean_var_a
## 1: cg14501977 10 -0.6653107 0.007728417 FALSE 0.01745930
## 2: cg02011981 10 -0.5970260 0.012028332 FALSE 0.03369357
## 3: cg25737283 10 -0.4803594 0.009757117 FALSE 0.04227059
## 4: cg11150667 10 -0.2643837 0.003424957 FALSE 0.04904385
## 5: cg07644184 10 -0.4876997 0.008267879 FALSE 0.03479852
## train_mean_aupr validation_mean_aupr cpg_score train_rank test_aupr
## 1: 1.0000000 1.000 0.001396744 1 0.9808673
## 2: 1.0000000 1.000 0.002695486 2 0.8743197
## 3: 0.9556481 1.000 0.003825165 3 1.0000000
## 4: 0.7621032 0.925 0.007052477 4 1.0000000
## 5: 0.5982708 0.775 0.009051174 5 0.6837868
##
## $glia$elapsed_time
## Time difference of 0.5351162 secs
##
##
## $blood_cells
## $blood_cells$train_res
## $blood_cells$train_res$fold_id
## # A tibble: 4,090 × 3
## Row Data Fold
## <int> <chr> <chr>
## 1 1 Analysis Fold01
## 2 1 Analysis Fold02
## 3 1 Analysis Fold03
## 4 1 Analysis Fold04
## 5 1 Analysis Fold05
## 6 1 Analysis Fold06
## 7 1 Analysis Fold08
## 8 1 Analysis Fold09
## 9 1 Analysis Fold10
## 10 2 Analysis Fold01
## # ℹ 4,080 more rows
##
## $blood_cells$train_res$train_summary
## id stat_origin mean_aupr mean_var_a fold_presence
## 1: cg02522196 train_aupr 0.9989550 0.05494539 10
## 2: cg02522196 validation_aupr 0.9994294 0.05494539 10
## 3: cg04785083 train_aupr 0.9996123 0.02253060 10
## 4: cg04785083 validation_aupr 0.9993818 0.02253060 10
## 5: cg05051606 train_aupr 0.9705120 0.07597889 10
## 6: cg05051606 validation_aupr 0.9819607 0.07597889 10
## 7: cg14286208 train_aupr 0.9985731 0.06884537 10
## 8: cg14286208 validation_aupr 0.9979712 0.06884537 10
## 9: cg18993949 train_aupr 0.9982355 0.07246407 10
## 10: cg18993949 validation_aupr 0.9983854 0.07246407 10
##
## $blood_cells$train_res$dt_dmsv
## id diff_means sum_variance pred_type
## 1: cg02522196 -0.7380026 0.02991550 FALSE
## 2: cg04785083 -0.8168825 0.01502010 FALSE
## 3: cg05051606 0.6092552 0.02819070 TRUE
## 4: cg14286208 0.6353021 0.02777695 TRUE
## 5: cg18993949 0.4876390 0.01722916 TRUE
##
## $blood_cells$train_res$train_results
## id fold_presence diff_means sum_variance pred_type mean_var_a
## 1: cg04785083 10 -0.8168825 0.01502010 FALSE 0.02253060
## 2: cg02522196 10 -0.7380026 0.02991550 FALSE 0.05494539
## 3: cg14286208 10 0.6353021 0.02777695 TRUE 0.06884537
## 4: cg18993949 10 0.4876390 0.01722916 TRUE 0.07246407
## 5: cg05051606 10 0.6092552 0.02819070 TRUE 0.07597889
## train_aupr validation_aupr cpg_score train_rank
## 1: 0.9996123 0.9993818 0.001812508 1
## 2: 0.9989550 0.9994294 0.004411787 2
## 3: 0.9985731 0.9979712 0.005542186 3
## 4: 0.9982355 0.9983854 0.005830916 4
## 5: 0.9705120 0.9819607 0.006553584 5
##
##
## $blood_cells$test_perf
## id fold_presence diff_means sum_variance pred_type mean_var_a
## 1: cg04785083 10 -0.8168825 0.01502010 FALSE 0.02253060
## 2: cg02522196 10 -0.7380026 0.02991550 FALSE 0.05494539
## 3: cg14286208 10 0.6353021 0.02777695 TRUE 0.06884537
## 4: cg18993949 10 0.4876390 0.01722916 TRUE 0.07246407
## 5: cg05051606 10 0.6092552 0.02819070 TRUE 0.07597889
## train_mean_aupr validation_mean_aupr cpg_score train_rank test_aupr
## 1: 0.9996123 0.9993818 0.001812508 1 0.9839162
## 2: 0.9989550 0.9994294 0.004411787 2 0.9596942
## 3: 0.9985731 0.9979712 0.005542186 3 0.7353430
## 4: 0.9982355 0.9983854 0.005830916 4 0.7801676
## 5: 0.9705120 0.9819607 0.006553584 5 0.8978199
##
## $blood_cells$elapsed_time
## Time difference of 0.412195 secs
##
##
## $fibroblasts
## $fibroblasts$train_res
## $fibroblasts$train_res$fold_id
## # A tibble: 4,090 × 3
## Row Data Fold
## <int> <chr> <chr>
## 1 1 Analysis Fold01
## 2 1 Analysis Fold02
## 3 1 Analysis Fold04
## 4 1 Analysis Fold05
## 5 1 Analysis Fold06
## 6 1 Analysis Fold07
## 7 1 Analysis Fold08
## 8 1 Analysis Fold09
## 9 1 Analysis Fold10
## 10 2 Analysis Fold01
## # ℹ 4,080 more rows
##
## $fibroblasts$train_res$train_summary
## id stat_origin mean_aupr mean_var_a fold_presence
## 1: cg02837162 train_aupr 0.6718360 0.3429565 5
## 2: cg02837162 validation_aupr 0.6244753 0.3429565 5
## 3: cg02907837 train_aupr 0.6822593 0.2570682 10
## 4: cg02907837 validation_aupr 0.7013197 0.2570682 10
## 5: cg03369247 train_aupr 0.8324525 0.2781751 10
## 6: cg03369247 validation_aupr 0.8561180 0.2781751 10
## 7: cg03509193 train_aupr 0.6585173 0.3324427 9
## 8: cg03509193 validation_aupr 0.7008693 0.3324427 9
## 9: cg26165286 train_aupr 0.6508256 0.3180929 6
## 10: cg26165286 validation_aupr 0.6143620 0.3180929 6
##
## $fibroblasts$train_res$dt_dmsv
## id diff_means sum_variance pred_type
## 1: cg02837162 -0.3433228 0.04093078 FALSE
## 2: cg02907837 -0.4095691 0.04309653 FALSE
## 3: cg03369247 -0.5998845 0.10002811 FALSE
## 4: cg03509193 -0.3343467 0.03717846 FALSE
## 5: cg26165286 -0.5503741 0.10177788 FALSE
##
## $fibroblasts$train_res$train_results
## id fold_presence diff_means sum_variance pred_type mean_var_a
## 1: cg03369247 10 -0.5998845 0.10002811 FALSE 0.2781751
## 2: cg02907837 10 -0.4095691 0.04309653 FALSE 0.2570682
## 3: cg03509193 9 -0.3343467 0.03717846 FALSE 0.3324427
## 4: cg26165286 6 -0.5503741 0.10177788 FALSE 0.3180929
## 5: cg02837162 5 -0.3433228 0.04093078 FALSE 0.3429565
## train_aupr validation_aupr cpg_score train_rank
## 1: 0.8324525 0.8561180 0.02536830 1
## 2: 0.6822593 0.7013197 0.02672967 2
## 3: 0.6585173 0.7008693 0.03666839 3
## 4: 0.6508256 0.6143620 0.05465925 4
## 5: 0.6718360 0.6244753 0.06894682 5
##
##
## $fibroblasts$test_perf
## id fold_presence diff_means sum_variance pred_type mean_var_a
## 1: cg03369247 10 -0.5998845 0.10002811 FALSE 0.2781751
## 2: cg02907837 10 -0.4095691 0.04309653 FALSE 0.2570682
## 3: cg03509193 9 -0.3343467 0.03717846 FALSE 0.3324427
## 4: cg26165286 6 -0.5503741 0.10177788 FALSE 0.3180929
## 5: cg02837162 5 -0.3433228 0.04093078 FALSE 0.3429565
## train_mean_aupr validation_mean_aupr cpg_score train_rank test_aupr
## 1: 0.8324525 0.8561180 0.02536830 1 0.7548379
## 2: 0.6822593 0.7013197 0.02672967 2 0.7377733
## 3: 0.6585173 0.7008693 0.03666839 3 0.6241444
## 4: 0.6508256 0.6143620 0.05465925 4 0.9547866
## 5: 0.6718360 0.6244753 0.06894682 5 0.4816497
##
## $fibroblasts$elapsed_time
## Time difference of 0.4029391 secs
# adjust target names to match signature names绘制 CimpleG CpG signature
# check generated signatures
plt <- signature_plot(
cimpleg_result,
train_data,
train_targets,
sample_id_column = "gsm",
true_label_column = "cell_type"
)
print(plt$plot)
均值之差与方差和(dmsv)图
基本的绘图
plt <- diffmeans_sumvariance_plot(
data = train_data,
target_vector = train_targets$neurons == 1
)
print(plt)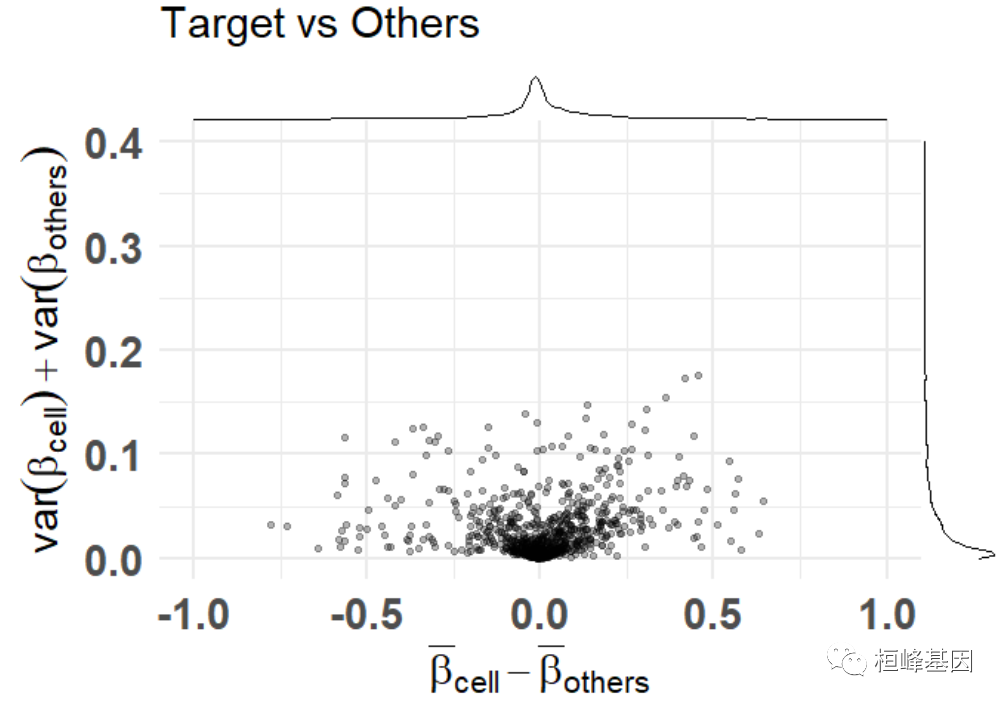
添加颜色,突出显示选定的功能
df_dmeansvar <- compute_diffmeans_sumvar(
data = train_data,
target_vector = train_targets$neurons == 1
)
parab_param <- .7
df_dmeansvar$is_selected <- select_features(
x = df_dmeansvar$diff_means,
y = df_dmeansvar$sum_variance,
a = parab_param
)
plt <- diffmeans_sumvariance_plot(
data = df_dmeansvar,
label_var1 = "Neurons",
color_all_points = "purple",
threshold_func = function(x, a) (a * x) ^ 2,
is_feature_selected_col = "is_selected",
func_factor = parab_param
)
print(plt)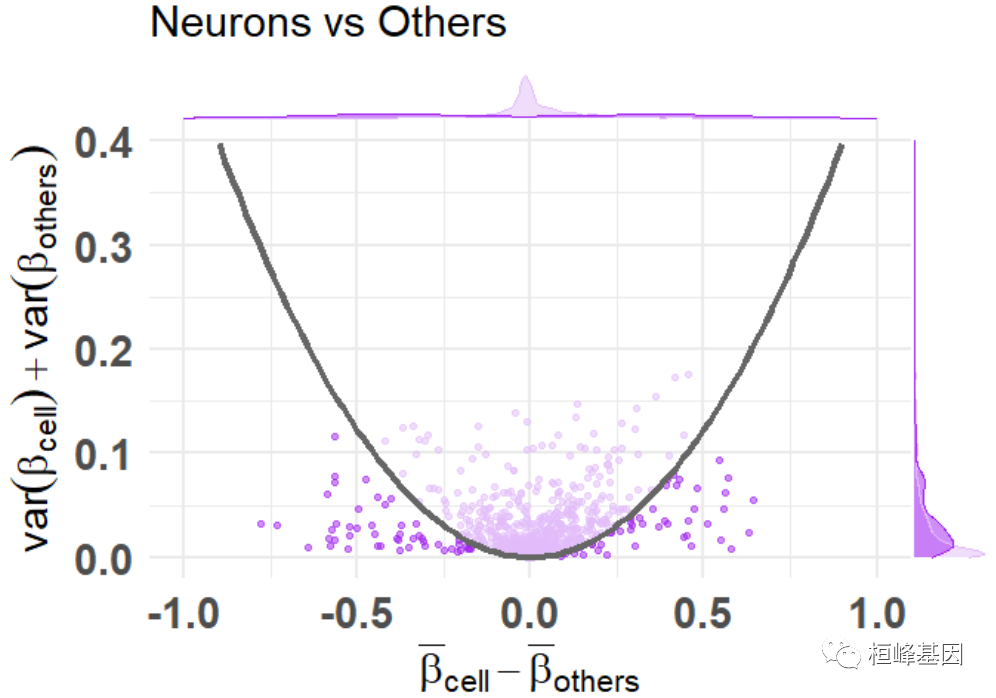
标记特征基因
plt <- diffmeans_sumvariance_plot(
data = df_dmeansvar,
feats_to_highlight = cimpleg_result$signatures
)
print(plt)
绘制反卷积图
最小的例子只有4个signature
deconv_result <- run_deconvolution(
cpg_obj = cimpleg_result,
new_data = test_data
)
plt <- deconvolution_barplot(
deconvoluted_data = deconv_result,
meta_data = test_targets,
sample_id = "gsm",
true_label = "cell_type"
)
print(plt$plot)
更高级的例子
这个例子更高级一些。首先,创建额外的反卷积结果,以便我们可以比较,使用CimpleG创建另外两个模型。一种只使用高甲基化的特征,另一种每个特征使用3个CpGs,而不是一个。
set.seed(42)
cimpleg_hyper <- CimpleG(
train_data = train_data,
train_targets = train_targets,
test_data = test_data,
test_targets = test_targets,
method = "CimpleG",
pred_type = "hyper",
target_columns = c(
"neurons",
"glia",
"blood_cells",
"fibroblasts"
)
)
## Training for target 'neurons' with 'CimpleG' has finished.: 0.48 sec elapsed
## Training for target 'glia' with 'CimpleG' has finished.: 0.28 sec elapsed
## Training for target 'blood_cells' with 'CimpleG' has finished.: 0.33 sec elapsed
## Training for target 'fibroblasts' with 'CimpleG' has finished.: 0.28 sec elapsed
deconv_hyper <- run_deconvolution(
cpg_obj = cimpleg_hyper,
new_data = test_data
)
set.seed(42)
cimpleg_3sigs <- CimpleG(
train_data = train_data,
train_targets = train_targets,
test_data = test_data,
test_targets = test_targets,
method = "CimpleG",
n_sigs = 3,
target_columns = c(
"neurons",
"glia",
"blood_cells",
"fibroblasts"
)
)
## Training for target 'neurons' with 'CimpleG' has finished.: 0.38 sec elapsed
## Training for target 'glia' with 'CimpleG' has finished.: 0.36 sec elapsed
## Training for target 'blood_cells' with 'CimpleG' has finished.: 0.37 sec elapsed
## Training for target 'fibroblasts' with 'CimpleG' has finished.: 0.5 sec elapsed
deconv_3sigs <- run_deconvolution(
cpg_obj = cimpleg_3sigs,
new_data = test_data
)让我们也创建一些假的真值,以便我们可以比较所有的结果。记住这只是一个例子,结果本身是没有意义的!
deconv_3sigs$prop_3sigs <- deconv_3sigs$proportion
deconv_hyper$prop_hyper <- deconv_hyper$proportion
deconv_result$prop_cimpleg <- deconv_result$proportion
dummy_deconvolution_data <-
deconv_result |>
dplyr::mutate(true_vals = proportion + runif(nrow(deconv_result), min=-0.1,max=0.1)) |>
dplyr::select(cell_type,sample_id,prop_cimpleg,true_vals) |>
dplyr::left_join(deconv_hyper |> dplyr::select(-proportion), by=c("sample_id","cell_type")) |>
dplyr::left_join(deconv_3sigs |> dplyr::select(-proportion), by=c("sample_id","cell_type")) |>
dplyr::mutate_if(is.numeric, function(x){ifelse(x<0,0,x)}) |>
dplyr::mutate_if(is.numeric, function(x){ifelse(x>1,1,x)}) |>
tibble::as_tibble()现在让我们利用一些用来比较反卷积结果的绘图函数,我们可以检查真实值与预测值的比较。
install.packages("broom")
## 程序包'broom'打开成功,MD5和检查也通过
##
## 下载的二进制程序包在
## C:\Users\Lenovo\AppData\Local\Temp\RtmpOonYSV\downloaded_packages里
scatter_plts <- CimpleG:::deconv_pred_obs_plot(
deconv_df = dummy_deconvolution_data,
true_values_col = "true_vals",
predicted_cols = c("prop_cimpleg","prop_hyper","prop_3sigs"),
sample_id_col = "sample_id",
group_col= "cell_type"
)
scatter_panel <- scatter_plts |> patchwork::wrap_plots(ncol=1)
print(scatter_panel)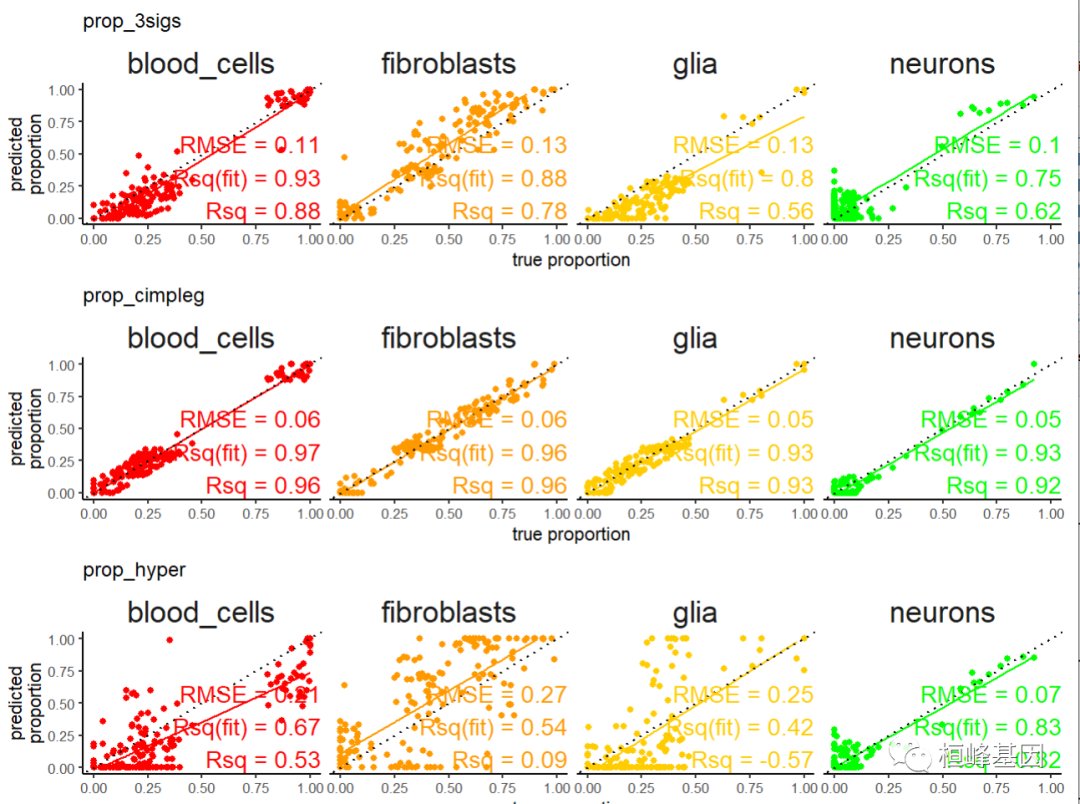
现在,更有趣的是,我们可以详细地看到并对用于评估反卷积结果的一个措施进行排序。
rank_plts <- CimpleG:::deconv_ranking_plot(
deconv_df = dummy_deconvolution_data,
true_values_col = "true_vals",
predicted_cols = c("prop_cimpleg","prop_hyper","prop_3sigs"),
sample_id_col = "sample_id",
group_col= "cell_type",
metrics = "rmse"
)
rank_panel <- list(rank_plts$perf_boxplt[[1]],rank_plts$nemenyi_plt[[1]]) |> patchwork::wrap_plots()
print(rank_panel)
Reference
Maié T, Schmidt M, Erz M, Wagner W, G Costa I. CimpleG: finding simple CpG methylation signatures. Genome Biol. 2023 Jul 10;24(1):161. doi: 10.1186/s13059-023-03000-0. PMID: 37430364; PMCID: PMC10332104.
桓峰基因,铸造成功的您!
未来桓峰基因公众号将不间断的推出单细胞系列生信分析教程,
敬请期待!!
桓峰基因官网正式上线,请大家多多关注,还有很多不足之处,大家多多指正!
http://www.kyohogene.com/
桓峰基因和投必得合作,文章润色优惠85折,需要文章润色的老师可以直接到网站输入领取桓峰基因专属优惠券码:KYOHOGENE,然后上传,付款时选择桓峰基因优惠券即可享受85折优惠哦!https://www.topeditsci.com/