虽然plink2.0已经存在好久了,但是一直用的都是plink1.9,因为语法熟悉。更主要是plink2.0语法变动太大,害怕步子迈得太大了……
今天看一下plink2.0的读入和输出数据常用参数,
plink2.0用是不会用的,2022年都不会用!!!但是碰到bgen,pgen数据进行转化为bed,bim,fam文件,然后用plink1.9使用的想法还是有的,而且很大!!!
本篇目的:使用plink2.0软件将下面格式随便输入、输出
- plink1.9的ped和map数据,不如:a.ped, a.map
- plink1.9的bed和bim和fam数据,比如:a.bim, a.bed, a.fam
- plink2.0的bgen和sample数据,比如:a.bgen, a.sample
- plink2.0的pgen和bim和fam数据,比如a.pgen,a.fam,a.bim
-
- vcf数据,比如a.vcf
1,plink2.0的提升
plink2.0主要是从以下几个方面,相对于plink1.9有较大的提升:
-
1,保留参考等位基因的信息,比如vcf格式的数据,不要添加参数 --keep-allele-order。这样vcf变为plink,plink变为vcf就可以不用指定ref和alt了,切换无障碍!
-
2,新的.pgen文件,结合SNPack-style的压缩,可以节约80%的文件大小。比如1000个Genomes,比压缩的gzip文件小70%,且不丢失任何信息。压缩文件空间更小,速度更快。
-
3,旧版的二进制文件(bed,bim和fam)文件,plink2.0依旧支持,输出文件包括两种:–make-bpgen 和 --make-bpfile文件。可以支持plink1.9的文件格式,无论是map和ped数据,还是bed,bim和fam格式。
-
4,分析模块,进行了优化。标准的logistic回归分析失败产生NA或者无意义的结果,–glm比plink1.9的–linear速度提升1000倍。尤其是填充的剂量效应的基因型值(比如0.2,1.8这样的非整数型数据)。PCA分析汇总,增加了参数
PCA approx,当样本超过1万,这个参数可以不影响精度(影响不到1%),大大提升计算效率。样本量支持得更多,处理速度更快!
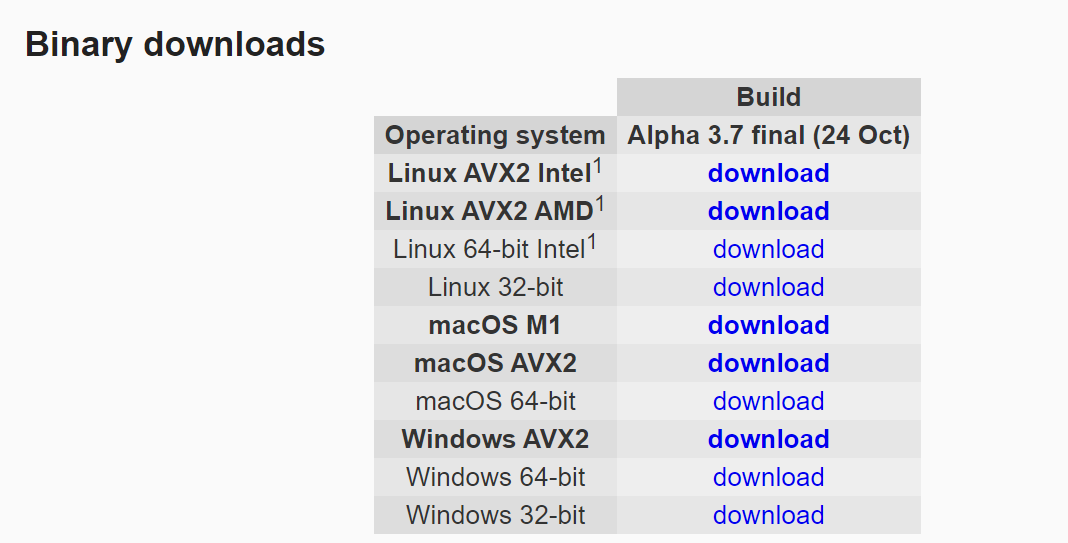
2,plink2.0 安装
plink2.0 网站:https://www.cog-genomics.org/plink/2.0/
二进制文件,直接执行,支持:
- Intel
- AMD
- 苹果M1
建议:plink1.9简写为plink,plink2.0 简写为plink2
3,plink帮助文档
可以通过官网查询具体参数:https://www.cog-genomics.org/plink/2.0/
也可以在命令行中调出帮助文档:
比如直接键入plink2,出现基础参数:
$ plink2
PLINK v2.00a3.7LM AVX2 Intel (24 Oct 2022) www.cog-genomics.org/plink/2.0/
(C) 2005-2022 Shaun Purcell, Christopher Chang GNU General Public License v3
plink2 <input flag(s)...> [command flag(s)...] [other flag(s)...]
plink2 --help [flag name(s)...]
Commands include --rm-dup list, --make-bpgen, --export, --freq, --geno-counts,
--sample-counts, --missing, --hardy, --het, --fst, --indep-pairwise, --ld,
--sample-diff, --make-king, --king-cutoff, --pmerge, --pgen-diff,
--write-samples, --write-snplist, --make-grm-list, --pca, --glm, --adjust-file,
--score, --variant-score, --genotyping-rate, --pgen-info, --validate, and
--zst-decompress.
"plink2 --help | more" describes all functions.
想查看一下–export的用法,可以看到主要功能:
- A,是0-1-2编码
- ped,是map和ped格式
- vcf,是vcf格式
- bgen-1.x,包括1.1, 1.2, 1.3,都是bgen格式
$ plink2 --export --help
PLINK v2.00a3.7LM AVX2 Intel (24 Oct 2022) www.cog-genomics.org/plink/2.0/
(C) 2005-2022 Shaun Purcell, Christopher Chang GNU General Public License v3
--help present, ignoring other flags.
--export <output format(s)...> [{01 | 12}] ['bgz'] ['id-delim='<char>]
['id-paste='<column set descriptor>] ['include-alt']
['omit-nonmale-y'] ['spaces'] ['vcf-dosage='<field>] ['ref-first']
['bits='<#>] ['sample-v2']
Create a new fileset with all filters applied. The following output
formats are supported:
(actually, only A, AD, Av, bcf, bgen-1.x, haps, hapslegend, ind-major-bed,
oxford, ped, tped, and vcf are implemented for now)
* '23': 23andMe 4-column format. This can only be used on a single
sample's data (--keep may be handy), and does not support
multicharacter allele codes.
* 'A': Sample-major additive (0/1/2) coding, suitable for loading from R.
If you need uncounted alleles to be named in the header line, add
the 'include-alt' modifier.
* 'AD': Sample-major additive (0/1/2) + dominant (het=1/hom=0) coding.
Also supports 'include-alt'.
* 'Av': Variant-major 0/1/2.
* 'beagle': Unphased per-autosome .dat and .map files, readable by early
BEAGLE versions.
* 'beagle-nomap': Single .beagle.dat file.
* 'bgen-1.x': Oxford-format .bgen + .sample. For v1.2/v1.3, sample
identifiers are stored in the .bgen (with id-delim and
id-paste settings applied), and default precision is 16-bit
(use the 'bits' modifier to reduce this).
* 'bimbam': Regular BIMBAM format.
* 'bimbam-1chr': BIMBAM format, with a two-column .pos.txt file. Does not
support multiple chromosomes.
* 'fastphase': Per-chromosome fastPHASE files, with
.chr-<chr #>.phase.inp filename extensions.
* 'fastphase-1chr': Single .phase.inp file. Does not support
multiple chromosomes.
* 'haps', 'hapslegend': Oxford-format .haps + .sample[ + .legend]. All
data must be biallelic and phased. When the 'bgz'
modifier is present, the .haps file is
block-gzipped.
* 'HV': Per-chromosome Haploview files, with .chr-<chr #>{.ped,.info}
filename extensions.
* 'HV-1chr': Single Haploview .ped + .info file pair. Does not support
multiple chromosomes.
* 'ind-major-bed': PLINK 1 sample-major .bed (+ .bim + .fam).
* 'lgen': PLINK 1 long-format (.lgen + .fam + .map), loadable with --lfile.
* 'lgen-ref': .lgen + .fam + .map + .ref, loadable with --lfile +
--reference.
* 'list': Single genotype-based list, up to 4 lines per variant. To omit
nonmale genotypes on the Y chromosome, add the 'omit-nonmale-y'
modifier.
* 'rlist': .rlist + .fam + .map fileset, where the .rlist file is a
genotype-based list which omits the most common genotype for
each variant. Also supports 'omit-nonmale-y'.
* 'oxford', 'oxford-v2': Oxford-format .gen + .sample. When the 'bgz'
modifier is present, the .gen file is
block-gzipped. 'oxford' requests the original
.gen file format with 5 leading columns
(understood by older PLINK builds); 'oxford-v2'
requests the current 6-leading-column flavor.
* 'ped', 'compound-genotypes': PLINK 1 sample-major (.ped + .map),
loadable with --pedmap.
* 'structure': Structure-format.
* 'tped': PLINK 1 variant-major (.tped + .tfam), loadable with --tfile.
* 'vcf', : VCF (default version 4.3). If PAR1 and PAR2 are present,
'vcf-4.2', they are automatically merged with chrX, with proper
'bcf', handling of chromosome codes and male ploidy.
'bcf-4.2' When the 'bgz' modifier is present, the VCF file is
block-gzipped. (This always happens with BCF output.)
The 'id-paste' modifier controls which .psam columns are
used to construct sample IDs (choices are maybefid, fid,
iid, maybesid, and sid; default is maybefid,iid,maybesid),
while the 'id-delim' modifier sets the character between the
ID pieces (default '_').
Genotypes are always exported. If you want to export a
sites-only VCF instead, see --make-pgen/--make-just-pvar's
'vcfheader' column set.
Dosages are not exported unless the 'vcf-dosage=' modifier
is present. The following six dosage export modes are
supported:
'GP': genotype posterior probabilities (v4.3 only).
'DS': Minimac3-style dosages, omitted for hardcalls.
'DS-force': Minimac3-style dosages, never omit.
'DS-only': Same as DS-force, except GT field is omitted.
'HDS': Minimac3-style phased dosages, omitted for hardcalls
and unphased calls. Also includes 'DS' output.
'HDS-force': Always report DS and HDS.
In addition,
* The '12' modifier causes alt1 alleles to be coded as '1' and ref alleles
to be coded as '2', while '01' maps alt1 -> 0 and ref -> 1.
* The 'spaces' modifier makes the output space-delimited instead of
tab-delimited, whenever both are permitted.
* For biallelic formats where it's unspecified whether the reference/major
allele should appear first or second, --export defaults to second for
compatibility with PLINK 1.9. Use 'ref-first' to change this.
(Note that this doesn't apply to the 'A', 'AD', and 'Av' formats; use
--export-allele to control which alleles are counted there.)
* 'sample-v2' exports .sample files according to the QCTOOLv2 rather than
the original specification. Only one ID column is exported ('id-paste'
and 'id-delim' settings apply), parental IDs are exported if present, and
category names are preserved rather than converted to positive integers.
--export-allele <file> : With --export A/AD/Av, count alleles named in the
file, instead of REF alleles.
4. plink2.0 笔记
4.1 读取plink的ped和map数据
plink2.0,没有–file这个参数了,变为了:--pedmap,也可以分开写,比如 --ped --map分别接ped和map数据。
plink2 --ped yuanshi.ped --map yuanshi.map
或者写为:
plink2 --pedmap yuanshi
默认输出文件:
plink2.log plink2.pgen plink2.psam plink2.pvar
- plink2.log,log日志,不用理会
- plink2.pgen,二进制文件,类似plink1.9的bim文件
- plink2.psam,个体和性别信息
- plink2.pvar,snp的信息,包括染色体、物理位置、名称、ref和alt
4.2 读取plink的bed,bim和fam数据
plink1.9的二进制数据是bed,bim和fam数据,plink2.0通过–bfile指定。
比如读取plink1.9的二进制文件,输出bgen格式:
plink2 --bfile a1 --export bgen-1.1 --out t1
日志:
$ plink2 --bfile a1 --export bgen-1.1 --out t1
PLINK v2.00a3.7LM AVX2 Intel (24 Oct 2022) www.cog-genomics.org/plink/2.0/
(C) 2005-2022 Shaun Purcell, Christopher Chang GNU General Public License v3
Logging to t1.log.
Options in effect:
--bfile a1
--export bgen-1.1
--out t1
Start time: Thu Dec 1 18:59:10 2022
1031523 MiB RAM detected; reserving 515761 MiB for main workspace.
Using up to 96 threads (change this with --threads).
86 samples (0 females, 0 males, 86 ambiguous; 86 founders) loaded from a1.fam.
50904 variants loaded from a1.bim.
Note: No phenotype data present.
Writing t1.bgen ... done.
Writing t1.sample ... done.
4.3 读取bgen和sample格式
导出ped和map的格式:
plink2 --bgen t1.bgen 'ref-last' --sample t1.sample --export ped --out x1
注意:bgen和sample是一对数据,读取时,要分开指定,–bgen, --sample,同时要指定‘ref-last’,即后面的是ref,前面的是alt,也可以修改。
4.4 读取vcf数据
导出ped和map数据
读取vcf,导出ped和map数据:需要定义--export ped
plink2 --vcf x2.vcf --export ped --out y1
导出bed和bim和fam数据
读取vcf数据,导出bim,bed和fam数据:需要定义--make-bed
plink2 --vcf x2.vcf --make-bed --out y2
导出bgen数据
读取vcf数据,导出bgen和sample数据,需要定义--export bgen-1.1
plink2 --vcf x2.vcf --export bgen-1.1 --out y3
上面就是我的总结,更多内容,欢迎关注我的公众号:育种数据分析之放飞自我












![[附源码]Python计算机毕业设计Django个性化产品服务管理系统论文](https://img-blog.csdnimg.cn/c4ee09901b214c28915167148dbdf5e0.png)