🎯要点
- 二维成像拥挤胶体粒子检测算法
- 粒子的局部结构和动力学分析
- 椭圆粒子成链动态过程定量分析算法
- 小颗粒的光散射和吸收
- 活跃物质模拟群体行为
- 提取粒子轨迹粘弹性,计算剪切模量
- 计算悬浮液球形粒子多体流体动力学
- 概率规划全息图跟踪和表征粒子位置、大小和折射率

Python粒子滤波器算法
为了简化,我们给出已经推导出的线性状态空间模型的粒子滤波器算法。粒子滤波器是针对以下状态空间模型推导出来的:
x
k
=
A
x
k
−
1
+
B
u
k
−
1
+
w
k
−
1
y
k
=
C
x
k
+
v
k
(
1
)
\begin{aligned} & x _k=A x _{k-1}+B u _{k-1}+ w _{k-1} \\ & y _k=C x _k+ v _k \end{aligned}\qquad(1)
xk=Axk−1+Buk−1+wk−1yk=Cxk+vk(1)
其中
x
k
∈
R
n
x _k \in R ^n
xk∈Rn 是离散时间步长
k
k
k 的状态向量,
u
k
−
1
∈
R
m
1
u _{k-1} \in R ^{m_1}
uk−1∈Rm1 是时间步长
k
−
1
k-1
k−1 的控制输入向量,
w
k
−
1
∈
R
m
2
w _{k-1} \in R ^{m_2}
wk−1∈Rm2 是时间步长
k
−
1
k-1
k−1 处的过程扰动向量(过程噪声向量),
y
k
∈
R
r
y _k \in R ^r
yk∈Rr 是时间步长
k
k
k 处观测到的输出矢量,
v
k
∈
R
r
v _k \in R ^r
vk∈Rr 是离散时间步长
k
k
k 处的测量噪声向量,
A
A
A,
B
B
B和
C
C
C是系统矩阵。
假设过程扰动向量
w
k
w _k
wk 服从正态分布,具有零均值和规定的协方差矩阵,即
w
k
∼
N
(
0
,
Q
)
(
2
)
w _k \sim N (0, Q)\qquad(2)
wk∼N(0,Q)(2)
其中
Q
Q
Q 是过程扰动向量的协方差矩阵。另外,假设测量噪声向量
v
k
v _k
vk 服从正态分布,具有零均值和规定的协方差矩阵,即
v
k
∼
N
(
0
,
R
)
(
3
)
v _k \sim N (0, R)\qquad(3)
vk∼N(0,R)(3)
其中
R
R
R 是测量噪声向量的协方差矩阵。状态转移密度是以下正态分布的密度
N
(
A
x
k
−
1
+
B
u
k
−
1
,
Q
)
(
4
)
N \left(A x _{k-1}+B u _{k-1}, Q\right)\qquad(4)
N(Axk−1+Buk−1,Q)(4)
此外,我们还证明了测量密度(测量概率密度函数),表示为
p
(
y
k
∣
x
k
)
p\left( y _k \mid x _k\right)
p(yk∣xk),是一个正态分布,其平均值为
C
x
k
C x _k
Cxk,协方差矩阵等于测量噪声向量
v
k
v _k
vk 的协方差矩阵。也就是说,测量密度是以下正态分布的密度
N
(
C
x
k
,
R
)
(
5
)
N \left(C x _k, R\right)\qquad(5)
N(Cxk,R)(5)
为了实现粒子滤波器,我们需要从状态转换概率 (4) 中抽取
x
k
x _k
xk 的样本。有两种方法可用于生成这些样本。第一种方法(我们在 Python 实现中使用)是从 (2) 中给出的分布中抽取过程扰动向量的随机样本。
在每个离散时间点
k
k
k,粒子滤波器计算以下一组粒子
{
(
x
k
(
i
)
,
w
k
(
i
)
)
∣
i
=
1
,
2
,
3
,
…
,
N
}
(
6
)
\left\{\left( x _k^{(i)}, w_k^{(i)}\right) \mid i=1,2,3, \ldots, N\right\}\qquad(6)
{(xk(i),wk(i))∣i=1,2,3,…,N}(6)
索引为
i
i
i 的粒子由元组
(
x
k
(
i
)
,
w
k
(
i
)
)
\left( x _k^{(i)}, w_k^{(i)}\right)
(xk(i),wk(i)) 组成,其中
x
k
(
i
)
x _k^{(i)}
xk(i) 是状态样本,
w
k
(
i
)
w_k^{(i)}
wk(i) 是重要性权重。粒子集近似后验密度
p
(
x
k
∣
y
0
:
k
,
u
0
:
k
−
1
)
p\left(x_k \mid y _{0: k}, u _{0: k-1}\right)
p(xk∣y0:k,u0:k−1),如下所示
p
(
x
k
∣
y
0
:
k
,
u
0
:
k
−
1
)
≈
∑
i
=
1
N
w
k
(
i
)
δ
(
x
k
−
x
k
(
i
)
)
(
7
)
p\left( x _k \mid y _{0: k}, u _{0: k-1}\right) \approx \sum_{i=1}^N w_k^{(i)} \delta\left( x _k- x _k^{(i)}\right)\qquad(7)
p(xk∣y0:k,u0:k−1)≈i=1∑Nwk(i)δ(xk−xk(i))(7)
粒子滤波算法的说明:对于初始粒子集
{
(
x
0
(
i
)
,
w
0
(
i
)
)
∣
i
=
1
,
2
,
3
,
…
,
N
}
\left\{\left( x _0^{(i)}, w_0^{(i)}\right) \mid i=1,2,3, \ldots, N\right\}
{(x0(i),w0(i))∣i=1,2,3,…,N}
Python过滤实现(片段)
import time
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import multivariate_normal
def systematicResampling(weightArray):
N=len(weightArray)
cValues=[]
cValues.append(weightArray[0])
for i in range(N-1):
cValues.append(cValues[i]+weightArray[i+1])
startingPoint=np.random.uniform(low=0.0, high=1/(N))
resampledIndex=[]
for j in range(N):
currentPoint=startingPoint+(1/N)*(j)
s=0
while (currentPoint>cValues[s]):
s=s+1
resampledIndex.append(s)
return resampledIndex
meanProcess=np.array([0,0])
covarianceProcess=np.array([[0.002, 0],[0, 0.002]])
meanNoise=np.array([0])
covarianceNoise=np.array([[0.001]])
processDistribution=multivariate_normal(mean=meanProcess,cov=covarianceProcess)
noiseDistribution=multivariate_normal(mean=meanNoise,cov=covarianceNoise)
m=5
ks=200
kd=30
Ac=np.array([[0,1],[-ks/m, -kd/m]])
Cc=np.array([[1,0]])
Bc=np.array([[0],[1/m]])
h=0.01
A=np.linalg.inv(np.eye(2)-h*Ac)
B=h*np.matmul(A,Bc)
C=Cc
simTime=1000
x0=np.array([[0.1],[0.01]])
stateDim,tmp11=x0.shape
controlInput=100*np.ones((1,simTime))
stateTrajectory=np.zeros(shape=(stateDim,simTime+1))
output=np.zeros(shape=(1,simTime))
stateTrajectory[:,[0]]=x0
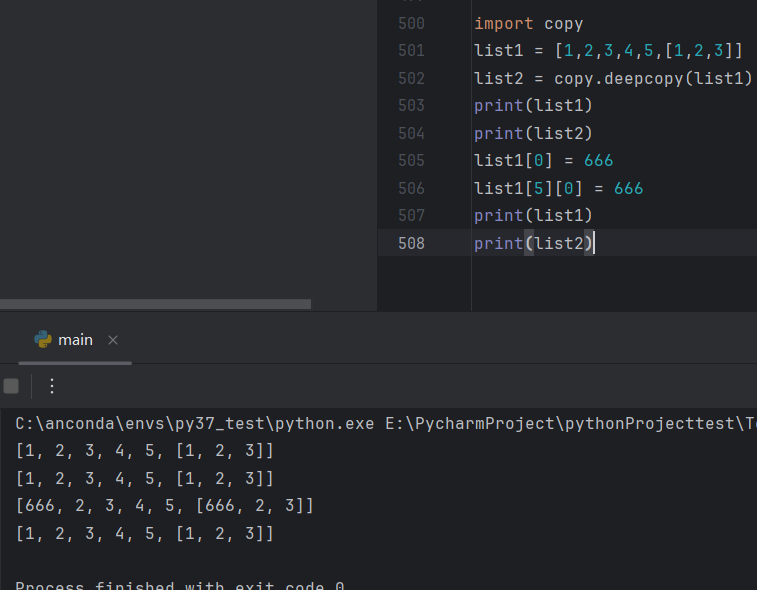
for i in range(simTime):
stateTrajectory[:,[i+1]]=np.matmul(A,stateTrajectory[:,[i]])+np.matmul(B,controlInput[:,[i]])+processDistribution.rvs(size=1).reshape(stateDim,1)
output[:,[i]]=np.matmul(C,stateTrajectory[:,[i]])+noiseDistribution.rvs(size=1).reshape(1,1)
x0Guess=x0+np.array([[0.7],[-0.6]])
pointsX, pointsY = np.mgrid[x0Guess[0,0]-0.8:x0Guess[0,0]+0.8:0.1, x0Guess[1,0]-0.5:x0Guess[1,0]+0.5:0.1]
xVec=pointsX.reshape((-1, 1), order="C")
yVec=pointsY.reshape((-1, 1), order="C")
states=np.hstack((xVec,yVec)).transpose()
dim1,numberParticle=states.shape
weights=(1/numberParticle)*np.ones((1,numberParticle))
numberIterations=1000
stateList=[]
stateList.append(states)
weightList=[]
weightList.append(weights)
for i in range(numberIterations):
controlInputBatch=controlInput[0,i]*np.ones((1,numberParticle))
newStates=np.matmul(A,states)+np.matmul(B,controlInputBatch)+processDistribution.rvs(size=numberParticle).transpose()
newWeights=np.zeros(shape=(1,numberParticle))
for j in range(numberParticle):
meanDis=np.matmul(C,newStates[:,[j]])
distributionO=multivariate_normal(mean=meanDis[0],cov=covarianceNoise)
newWeights[:,j]=distributionO.pdf(output[:,i])*weights[:,[j]]
weightsStandardized=newWeights/(newWeights.sum())
tmp1=[val**2 for val in weightsStandardized]
Neff=1/(np.array(tmp1).sum())
if Neff<(numberParticle//3):
resampledStateIndex=np.random.choice(np.arange(numberParticle), numberParticle, p=weightsStandardized[0,:])
newStates=newStates[:,resampledStateIndex]
weightsStandardized=(1/numberParticle)*np.ones((1,numberParticle))
states=newStates
weights=weightsStandardized
stateList.append(states)
weightList.append(weights)
meanStateSequence=np.zeros(shape=(stateDim,numberIterations))
for i in range(numberIterations):
meanState=np.zeros(shape=(stateDim,1))
for j in range(numberParticle):
meanState=meanState+weightList[i][:,j]*stateList[i][:,j].reshape(2,1)
meanStateSequence[:,[i]]=meanState