Title
题目
Labeled-to-unlabeled distribution alignment for partially-supervised multi-organ medical image segmentation
部分监督多器官医学图像分割中的标记与未标记分布对齐
01
文献速递介绍
多器官医学图像分割(Mo-MedISeg)是医学图像分析领域中一个基础但具有挑战性的研究任务。它涉及为给定图像的每个像素分配一个语义器官标签,例如“肝脏”、“肾脏”、“脾脏”或“胰腺”。任何未定义的器官区域都被视为属于“背景”类别(Cerrolaza等,2019)。随着深度图像处理技术的发展,例如卷积神经网络(CNNs)(He等,2016)和视觉变换器(Dosovitskiy等,2020;Zhang等,2020b,2024),Mo-MedISeg 引起了显著的研究关注,并被广泛应用于各种实际应用中,包括诊断干预(Kumar等,2019)和治疗规划(Chu等,2013),以及诸如计算机断层扫描(CT)图像(Gibson等,2018)和X射线图像(Gómez等,2020)等不同场景。
然而,训练一个完全监督的深度语义图像分割模型是具有挑战性的,因为这通常需要大量的像素级标注样本(Wei等,2016;Zhang等,2020a)。这一挑战在Mo-MedISeg任务中更加艰巨,因为获取精确且密集的多器官注释既繁琐又耗时,还需要稀缺的专家知识。因此,大多数基准公共数据集,如LiTS(Bilic等,2023)和KiTS(Heller等,2019),只提供一个器官的注释,而与任务无关的其他器官则被标记为背景。与完全标注的多器官数据集相比,部分注释的多器官数据集更容易获取。为了减轻收集完整注释的负担,部分监督学习(PSL)被用于从多个部分标注的数据集中学习Mo-MedISeg模型(Zhou等,2019;Zhang等,2021a;Liu和Zheng,2022)。在这种设置下,每个数据集为一个特定类别的器官提供标签,直到覆盖所有感兴趣的前景类别。该策略避免了对密集标注数据集的需求,并允许合并不同机构标注有不同类型器官的数据集,尤其是当不同医院关注不同器官时。
在Mo-MedISeg任务中使用PSL的一个关键挑战是如何在不复杂化多器官分割模型的情况下利用有限的标注像素和大量的未标注像素。每个部分标注的数据集都使用一个二进制图来为特定器官进行标注,指示某个像素是否属于目标器官。没有为其他前景器官和背景类别提供标签,导致每张图像的训练集同时包含标注和未标注像素。以往的方法通常仅从标注像素中学习(Dmitriev和Kaufman,2019;Zhou等,2019;Zhang等,2021a),或者将缺失的标签视为背景(Fang和Yan,2020;Shi等,2021)。然而,对未标注结构缺乏监督可能容易导致过拟合问题,而将未标注的解剖结构视为背景可能在没有完全注释数据集的先验信息的情况下误导和混淆Mo-MedISeg模型。尽管一些先进的方法(Huang等,2020;Feng等,2021;Liu和Zheng,2022)已经被提出来为未标注像素生成伪监督,这些方法通常只是采用自训练生成伪标签。它们使用标注像素训练教师模型,例如为每个数据集的多个单独单器官分割模型。然后,教师模型为每个部分标注数据集中的未注释器官生成伪标签,形成一个伪多器官数据集,用于训练学生多器官分割模型。学生模型的性能取决于伪标签的质量。不可靠的伪标签可能导致严重的确认偏差和性能下降(Chen等,2022a)。
Aastract
摘要
Partially-supervised multi-organ medical image segmentation aims to develop a unified semantic segmentationmodel by utilizing multiple partially-labeled datasets, with each dataset providing labels for a single classof organs. However, the limited availability of labeled foreground organs and the absence of supervision todistinguish unlabeled foreground organs from the background pose a significant challenge, which leads to adistribution mismatch between labeled and unlabeled pixels. Although existing pseudo-labeling methods can beemployed to learn from both labeled and unlabeled pixels, they are prone to performance degradation in thistask, as they rely on the assumption that labeled and unlabeled pixels have the same distribution. In this paper,to address the problem of distribution mismatch, we propose a labeled-to-unlabeled distribution alignment(LTUDA) framework that aligns feature distributions and enhances discriminative capability. Specifically,we introduce a cross-set data augmentation strategy, which performs region-level mixing between labeledand unlabeled organs to reduce distribution discrepancy and enrich the training set. Besides, we proposea prototype-based distribution alignment method that implicitly reduces intra-class variation and increasesthe separation between the unlabeled foreground and background. This can be achieved by encouragingconsistency between the outputs of two prototype classifiers and a linear classifier. Extensive experimentalresults on the AbdomenCT-1K dataset and a union of four benchmark datasets (including LiTS, MSD-Spleen,KiTS, and NIH82) demonstrate that our method outperforms the state-of-the-art partially-supervised methodsby a considerable margin, and even surpasses the fully-supervised methods.
部分监督多器官医学图像分割旨在通过利用多个部分标记的数据集来开发统一的语义分割模型,每个数据集为单个器官类别提供标签。然而,标记前景器官的可用性有限,且缺乏监督来区分未标记前景器官与背景,这给任务带来了重大挑战,导致标记和未标记像素之间的分布不匹配。尽管现有的伪标记方法可以用于同时学习标记和未标记像素,但由于它们依赖于标记和未标记像素具有相同分布的假设,因此在此任务中容易导致性能下降。为了解决分布不匹配的问题,我们提出了一种标记到未标记分布对齐(LTUDA)框架,该框架对齐特征分布并增强辨别能力。具体而言,我们引入了一种跨集数据增强策略,通过在标记和未标记器官之间进行区域级混合来减少分布差异并丰富训练集。此外,我们提出了一种基于原型的分布对齐方法,隐式减少类内变异性,并增加未标记前景与背景之间的分离。这可以通过鼓励两个原型分类器和一个线性分类器输出之间的一致性来实现。在AbdomenCT-1K数据集和四个基准数据集的联合(包括LiTS、MSD-Spleen、KiTS和NIH82)上进行的广泛实验结果表明,我们的方法在很大程度上超过了最先进的部分监督方法,甚至超越了完全监督的方法。
Method
方法
3.1. Preliminaries
Given a union of partially annotated datasets {𝐷1 , 𝐷2 , …, 𝐷𝐶} (𝐷𝑐= {(𝑋, 𝑌 𝑙 ) ∣ 𝑐 = 1,2, .. ., 𝐶}), our goal is to train an end-to-endsegmentation network that can simultaneously segment 𝐶 organs. 𝑋denotes the images in the 𝑐th dataset. The corresponding partial label𝑌* 𝑙 comprises labeled foreground organ pixels, unlabeled foregroundorgan pixels, and unlabeled background pixels. In 𝑌 𝑙 , only organ 𝑐has true class labels, and other organs are unknown. The multi-organsegmentation network needs to assign each pixel of an input image aunique semantic label of 𝑐 ∈ {0, 1, …, 𝐶}, where 𝐶 denotes the classsize.CNN-based Mo-MedISeg networks are commonly trained by multiclass cross-entropy loss L𝐶𝐸 (𝑥, 𝑦) = − ∑𝐶**𝑐=0 𝑦𝑐 log 𝑝𝑐 , where 𝑦𝑐 is theone-hot ground truth corresponding to class 𝑐. However, this is notsuitable for partially-supervised segmentation. Given the absence ofan annotated background class in this scenario, we learn class-specificforeground maps, and transform the multi-classification task into multiple binary classification tasks (one vs. the rest). Given an image fromany partially labeled dataset 𝐷**𝑖 as input, the segmentation networkgenerates class-wise foreground maps 𝑝 = {𝑝𝑐 } 𝐶 𝑐=1, which are normalized using a sigmoid. Partial binary cross-entropy loss L𝑝𝐵𝐶𝐸 is adoptedto train the segmentation network, which is defined as follows:
L𝑝𝐵𝐶𝐸 (𝑥, 𝑦) =𝐶∑𝑐=1I𝑦𝑐≠−1,
(1)where 𝑦𝑐 and 𝑝𝑐 represent the ground truth and predicted probabilitymap belonging to class c, respectively. For the labeled category 𝑖, thevalue of 𝑦𝑐 is 0 or 1, indicating whether or not the pixel belongs tocategory 𝑖; for the unlabeled categories, the value of 𝑦𝑐 is −1. L𝑝𝐵𝐶𝐸only supervises the segmentation map corresponding to the labeledcategory. For example, the LiTs dataset provides partial labels for theliver category. Therefore, for some other categories without labels, thisloss function will not back-propagate the gradients.The segmentation network only outputs foreground maps, so weneed to assign each pixel its unique class ̂𝑦 from the label space{0, 1, …, 𝐶} during inference. Inspired by methods for detecting anomalous samples (Hendrycks and Gimpel, 2016), we adopt a linear
threshold-based classifier to obtain multi-class segmentation maps. Asshown in Eq. (2), if the probabilities of all foreground classes of a pixelare below a certain threshold 𝜏, we consider the pixel to belong to thebackground class. Otherwise, the pixel is assigned the class with thehighest foreground probability.̂𝑦 = { arg max𝑐∈{1,…,𝐶}𝑝𝑐 , if max𝑐∈{1,…,𝐶}𝑝*𝑐 ≥ 𝜏,0 (background class)
,otherwise.(2)Although ignoring unannotated categories is an intuitive approach, itmay lead to sub-optimal performance as only a fraction of the dataare utilized. Fig. 1(a) shows the feature visualization of the baselinemodel which only learns from the labeled pixels while ignoring theunlabeled organs. We can observe that the unlabeled prototypes deviatefrom the labeled prototype for the foreground categories, especiallyfor the pancreas and spleen. Furthermore, it is difficult to separate theunlabeled foreground from the background only by the threshold 𝜏, dueto the absence of any background supervision information. Therefore,directly minimizing L𝑝𝐵𝐶𝐸 on labeled pixels may result in the problemsof overfitting and confusion between the foreground organs and thebackground.
给定一个部分标注数据集的联合体 { 𝐷1, 𝐷2, …, 𝐷𝐶 } (𝐷𝑐 = { (𝑋, 𝑌 𝑙) | 𝑐 = 1, 2, …, 𝐶 }),我们的目标是训练一个端到端的分割网络,能够同时对 𝐶 个器官进行分割。𝑋 表示第 𝑐 个数据集中的图像。对应的部分标签 𝑌 𝑙 包括标记的前景器官像素、未标记的前景器官像素和未标记的背景像素。在 𝑌 𝑙 中,只有器官 𝑐 有真实的类别标签,其他器官的标签未知。多器官分割网络需要将输入图像的每个像素分配一个唯一的语义标签 𝑐 ∈ {0, 1, …, 𝐶},其中 𝐶 表示类别数量。基于卷积神经网络的 Mo-MedISeg 网络通常通过多类别交叉熵损失 L𝐶𝐸 (𝑥, 𝑦) = − ∑𝐶𝑐=0 𝑦𝑐 log 𝑝𝑐 进行训练,其中 𝑦𝑐 是与类别 𝑐 对应的一热编码真实值。然而,这不适用于部分监督分割。由于在这种情况下缺少标注的背景类别,我们学习特定类别的前景图,并将多分类任务转换为多个二分类任务(一个类别与其余类别的比较)。对于来自任何部分标注数据集 𝐷𝑖 的图像,分割网络生成类别特定的前景图 𝑝 = {𝑝**𝑐 } 𝐶 𝑐=1,这些图通过 sigmoid 函数进行归一化。采用部分二元交叉熵损失 L𝑝𝐵𝐶𝐸 来训练分割网络,其定义如下:
L𝑝𝐵𝐶𝐸 (𝑥, 𝑦) = 𝐶∑𝑐=1 I𝑦𝑐≠ −1
(1) 其中 𝑦**𝑐 和 𝑝**𝑐 分别表示类别 𝑐 的真实值和预测概率图。对于标记类别 𝑖,𝑦𝑐 的值为 0 或 1,指示该像素是否属于类别 𝑖;对于未标记类别,𝑦𝑐 的值为 −1。L𝑝𝐵𝐶𝐸 仅对与标记类别对应的分割图进行监督。例如,LiTs 数据集为肝脏类别提供部分标签。因此,对于其他没有标签的类别,该损失函数将不会反向传播梯度。分割网络仅输出前景图,因此在推理过程中我们需要为每个像素分配一个唯一的类别 ̂𝑦,其标签空间为 {0, 1, …, 𝐶}。受到异常样本检测方法的启发(Hendrycks 和 Gimpel, 2016),我们采用基于线性阈值的分类器来获得多类别分割图。如下所示,如果某个像素的所有前景类别的概率都低于某个阈值 𝜏,我们认为该像素属于背景类别。否则,该像素将被分配给具有最高前景概率的类别。
̂𝑦 = { arg max𝑐∈{1, …, 𝐶} 𝑝𝑐, 如果 max𝑐∈{1, …, 𝐶} 𝑝𝑐 ≥ 𝜏,0(背景类),否则。
(2) 尽管忽略未标注类别是一种直观的方法,但这可能导致次优性能,因为仅使用了部分数据。图 1(a) 显示了基线模型的特征可视化,该模型仅从标记像素中学习,同时忽略了未标记器官。我们可以观察到,未标记的原型与前景类别的标记原型偏离,特别是对于胰腺和脾脏。此外,由于缺乏任何背景监督信息,仅通过阈值 𝜏 难以将未标记的前景与背景分离。因此,直接最小化 L𝑝𝐵𝐶𝐸 在标记像素上可能导致过拟合和前景器官与背景之间的混淆问题。
Conclusion
结论
Partially-supervised segmentation is confronted with the challengeof feature distribution mismatch between the labeled and unlabeledpixels. To address this issue, this paper proposes a new frameworkcalled LTUDA for partially supervised multi-organ segmentation. Ourapproach comprises two key components. Firstly, we design a crossset data augmentation strategy that generates interpolated samplesbetween the labeled and unlabeled pixels, thereby reducing featurediscrepancy. Furthermore, we propose a prototype-based distributionalignment module to align the distributions of labeled and unlabeledpixels. This module eliminates confusion between unlabeled foregroundand background by encouraging consistency between labeled prototypeand unlabeled prototype classifier. Experimental results on both a toydataset and a large-scale partially labeled dataset demonstrate theeffectiveness of our method.Our proposed method has the potential to contribute to the development of foundation models. Recent foundation models (Liu et al., 2023;Ye et al., 2023) have focused on leveraging large-scale and diversepartially annotated datasets to segment different types of organs andtumors, thereby fully exploiting existing publicly available datasetsfrom different modalities (e.g., CT, MRI, PET). While these models haveachieved promising results, they typically do not leverage unlabeledpixels or consider the domain shift between partially-labeled datasetsfrom different institutions. This heterogeneity in training data leadsto a more pronounced distributional deviation between labeled andunlabeled pixels, posing a greater challenge. Our proposed labeled-tounlabeled distribution framework specifically addresses this challengeand thus can help unlock the potential of public datasets. Lookingahead, one promising future direction is to apply partially supervisedsegmentation methods to more realistic clinical scenarios (Yao et al.,2021). Our approach demonstrates the feasibility of training multiorgan segmentation models using small-sized partially labeled datasets.This convenience enables the customization and development of multiorgan segmentation models based on the clinical requirements of medical professionals. If the annotations provided by the public datasetslack specific anatomical structures of interest, experts would only needto provide annotations of a small number of missing classes to developmulti-organ segmentation models.
部分监督分割面临标记像素和未标记像素之间特征分布不匹配的挑战。为了解决这个问题,本文提出了一种新的框架,称为 LTUDA,用于部分监督的多脏器分割。我们的方法包括两个关键组件。首先,我们设计了一种跨集合数据增强策略,生成标记像素和未标记像素之间的插值样本,从而减少特征差异。此外,我们提出了基于原型的分布对齐模块,以对齐标记像素和未标记像素的分布。该模块通过鼓励标记原型与未标记原型分类器之间的一致性,消除了未标记前景与背景之间的混淆。我们在玩具数据集和大规模部分标记数据集上的实验结果证明了我们方法的有效性。
我们提出的方法有潜力为基础模型的发展做出贡献。最近的基础模型(Liu et al., 2023; Ye et al., 2023)专注于利用大规模和多样化的部分注释数据集对不同类型的器官和肿瘤进行分割,从而充分利用来自不同模态(例如 CT、MRI、PET)的现有公共数据集。尽管这些模型取得了令人鼓舞的结果,但它们通常不利用未标记像素或考虑来自不同机构的部分标记数据集之间的领域偏移。这种训练数据的异质性导致标记像素和未标记像素之间的分布偏差更加明显,从而带来更大的挑战。我们提出的标记到未标记分布框架专门针对这一挑战,从而帮助挖掘公共数据集的潜力。展望未来,一个有前景的方向是将部分监督分割方法应用于更真实的临床场景(Yao et al., 2021)。我们的方法展示了使用小规模部分标记数据集训练多脏器分割模型的可行性。这种便利性使得根据医疗专业人员的临床需求定制和开发多脏器分割模型成为可能。如果公共数据集提供的注释缺少特定感兴趣的解剖结构,专家只需提供少量缺失类别的注释即可开发多脏器分割模型。
Figure
图
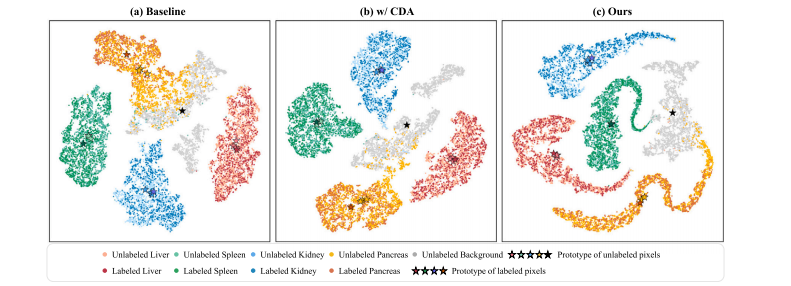
Fig. 1. Comparisons of t-SNE feature visualization on the toy dataset consisting of four partially-labeled sub-datasets. The feature distribution of labeled and unlabeled pixels fordifferent classes is visualized. For each foreground category, only one sub-dataset provides a labeled set, while the other three provide unlabeled sets. Since each sub-dataset doesnot provide the true label of the background, the background is completely unlabeled. We have superimposed the feature centers of the labeled set and unlabeled set, i.e., labeledprototypes and unlabeled prototypes, of each foreground category on the feature distribution. Additionally, we visualized the feature center of the background classes across allsubsets. (a) Baseline model (trained on labeled pixels). The labeled prototype and unlabeled prototypes of the foreground classes are not aligned. (b) Baseline model with cross-setdata augmentation (CDA). The CDA strategy effectively reduces the distributional discrepancy between labeled and unlabeled pixels for the foreground classes. (c) Our proposedmethod. The labeled prototype and unlabeled prototypes of each foreground class almost overlap
图 1. t-SNE 特征可视化在由四个部分标注的子数据集组成的玩具数据集上的比较。可视化了不同类别的标注和未标注像素的特征分布。对于每个前景类别,只有一个子数据集提供标注集,而其他三个子数据集提供未标注集。由于每个子数据集都没有提供背景的真实标签,背景是完全未标注的。我们在特征分布上叠加了每个前景类别的标注集和未标注集的特征中心,即标注原型和未标注原型。此外,我们可视化了所有子数据集中背景类别的特征中心。(a)基线模型(仅在标注像素上训练)。前景类别的标注原型和未标注原型未对齐。(b)带有跨集数据增强(CDA)的基线模型。CDA策略有效减少了前景类别中标注像素和未标注像素之间的分布差异。(c)我们提出的方法。每个前景类别的标注原型和未标注原型几乎完全重合。
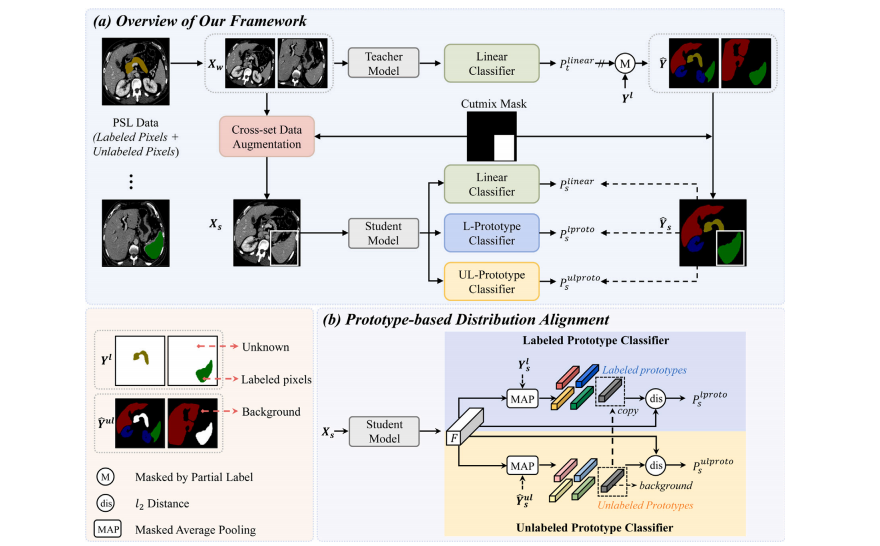
Fig. 2. (a) The overall framework of the proposed LTUDA method, which consists of cross-set data augmentation and prototype-based distribution alignment. (b) Details of theprototype-based distribution alignment module. Our method is built on the popular teacher–student framework and applies weak (rotation and scaling) and strong augmentation(cross-set region-level mixing) to the input images of the teacher and student models, respectively. The linear classifier refers to the linear threshold-based classifier describedin Eq. (2) of Section 3.1. Two prototype classifiers are introduced in the student model, and the predictions of the teacher model and partial labels are combined as pseudo-labelsto supervise the outputs of the three classifiers in the student model. The term ‘‘copy’’ denotes that the labeled prototype of the background class is set to be equal to the unlabeledprototype.
图 2. (a) 所提出的 LTUDA 方法的整体框架,包含跨集数据增强和基于原型的分布对齐。(b) 基于原型的分布对齐模块的详细信息。我们的方法基于流行的教师-学生框架,并分别对教师模型和学生模型的输入图像应用弱增强(旋转和缩放)和强增强(跨集区域级混合)。线性分类器指的是3.1节方程(2)中描述的线性阈值分类器。学生模型中引入了两个原型分类器,教师模型的预测和部分标签被结合为伪标签,用于监督学生模型中三个分类器的输出。术语“复制”表示背景类的标注原型设置为等于未标注原型。
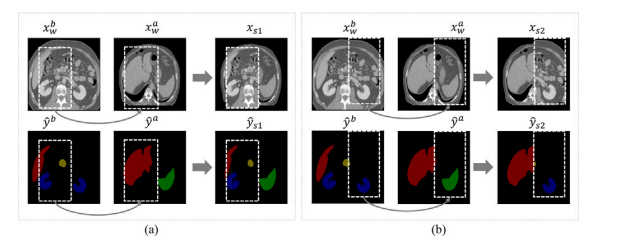
Fig. 3. Visualizations of differently strongly augmented images generated by CutMix. (a) and (b) paste the cropped patch from 𝑥 𝑏 𝑤 to the same position in 𝑥 𝑎 𝑤 . The box coordinates and sizes of (a) and (b) are different.
图 3. 使用 CutMix 生成的不同强增强图像的可视化。
(a) 和 (b) 将从 𝑥 𝑏 𝑤 裁剪的图像块粘贴到 𝑥 𝑎 𝑤 的相同位置。(a) 和 (b) 的裁剪框坐标和大小不同。
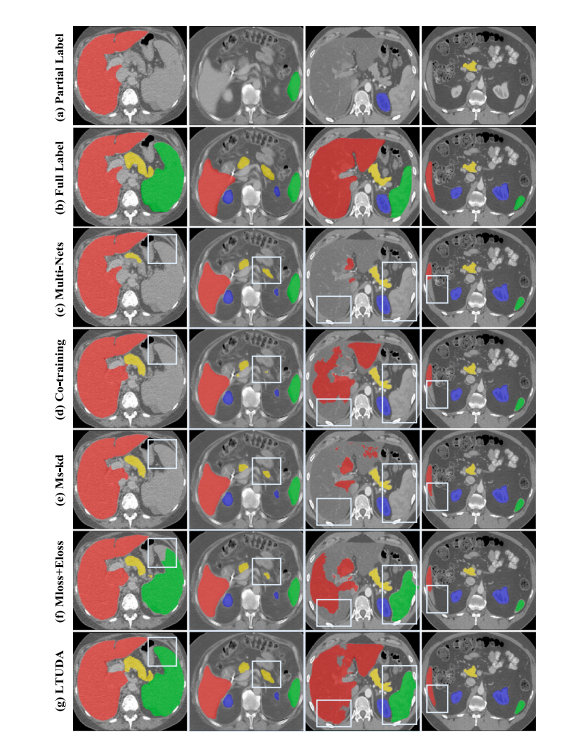
Fig. 4. Visualizations of LSPL. Examples are from the LiTS, MSD-Spleen, KiTS, andNIH82 datasets, respectively, arranged from left to right. (a) Single-organ annotationsoriginally provided by the four benchmark datasets. (b) Full annotations of four organs.(c) to (g) are the segmentation results of different methods. The white frame highlightsthe better predictions of our method.
图4. LSPL 的可视化示例。例子分别来自 LiTS、MSD-Spleen、KiTS 和 NIH82 数据集,按从左到右的顺序排列。(a)四个基准数据集原始提供的单脏器标注。(b)四个脏器的完整标注。(c)到(g)是不同方法的分割结果。白色框突出显示了我们方法的更好预测。
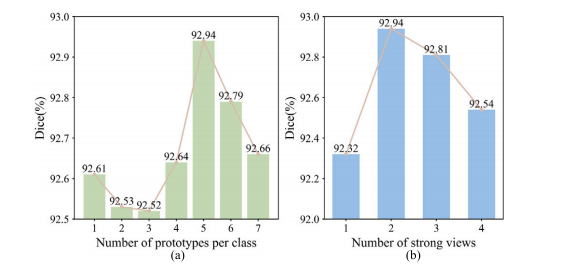
Fig. 5. Ablation results. (a) The number of prototypes per class. (b) The number ofstrong views.
图 5. 消融结果。(a) 每个类别的原型数量。(b) 强视图的数量。
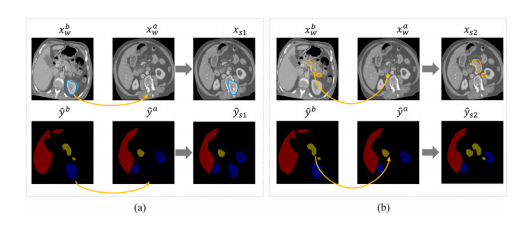
Fig. 6. Visualizations of differently strongly augmented images generated by CarveMix.(a) and (b) paste the cropped ROI from 𝑥 𝑏 𝑤 to the same position in 𝑥 𝑎 𝑤 . (a) and (b)crop the ROI of different foreground organs.
图 6. CarveMix 生成的不同强度增强图像的可视化。(a) 和 (b) 将从 𝑥 𝑏 𝑤 裁剪的 ROI 粘贴到 𝑥 𝑎 𝑤 的相同位置。(a) 和 (b) 裁剪不同前景器官的 ROI。
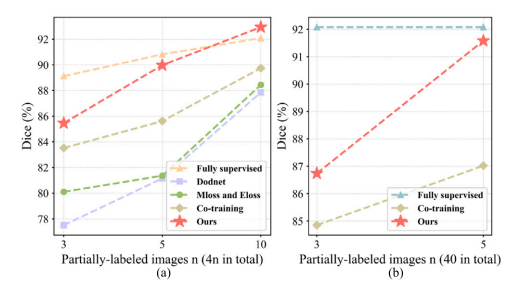
Fig. 7. (a) The performance of learning from partially labeled data under different labeled data sizes. (b) The performance of learning from partially labeled data and unlabeled data under different labeled data sizes.
图 7. (a) 在不同标记数据大小下从部分标记数据学习的性能。(b) 在不同标记数据大小下从部分标记数据和未标记数据学习的性能。
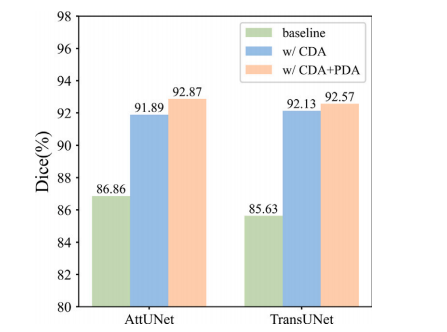
Fig. 8. Ablation results of different segmentation backbones.
图 8. 不同分割骨干网络的消融结果。
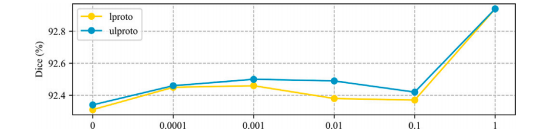
Fig. 9. Ablation study of weight parameters of L𝑙𝑝𝑟𝑜𝑡𝑜 and L𝑢𝑙𝑝𝑟𝑜𝑡𝑜.
图 9. L𝑙𝑝𝑟𝑜𝑡𝑜 和 L𝑢𝑙𝑝𝑟𝑜𝑡𝑜 权重参数的消融研究
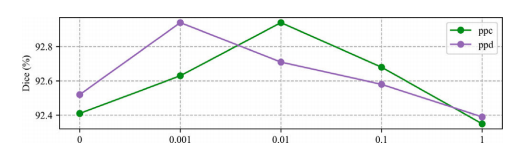
Fig. 10. Ablation study of weight parameters of L𝑝𝑝𝑑 and L𝑝𝑝𝑐 .
图 10. L𝑝𝑝𝑑 和 L𝑝𝑝𝑐 权重参数的消融研究。
Table
表
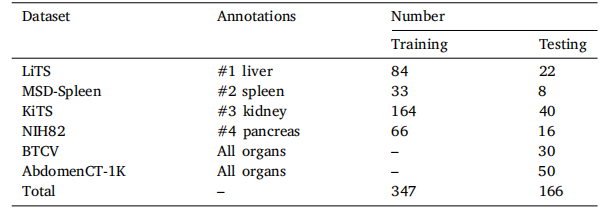
Table 1A brief description of the large-scale partially-labeled dataset.
表1 大规模部分标记数据集的简要描述。
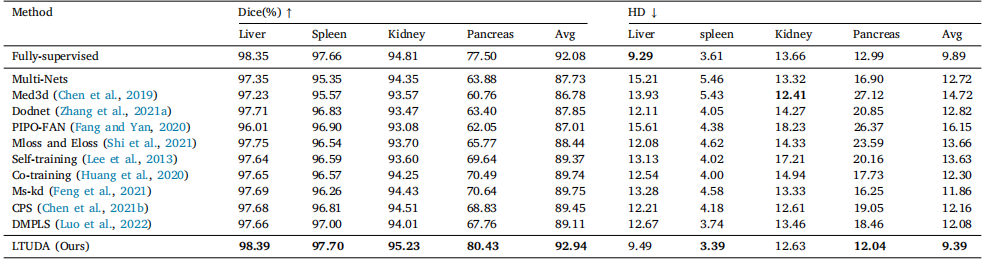
Table 2Quantitative results of partially-supervised multi-organ segmentation on a toy dataset.
表2 在玩具数据集上进行部分监督多脏器分割的定量结果。
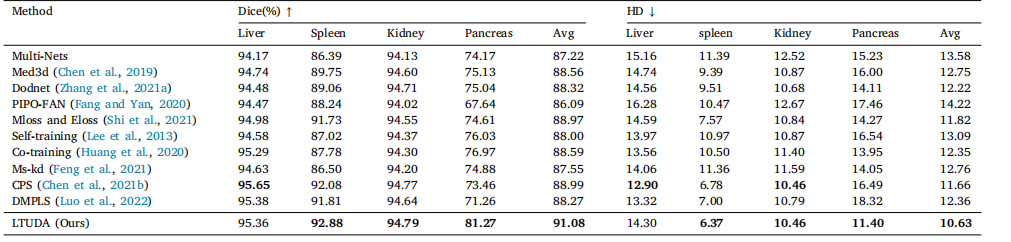
Table 3Quantitative results of partially-supervised multi-organ segmentation on four partially labeled datasets (LSPL dataset).
表3 在四个部分标记数据集(LSPL 数据集)上进行部分监督多脏器分割的定量结果。
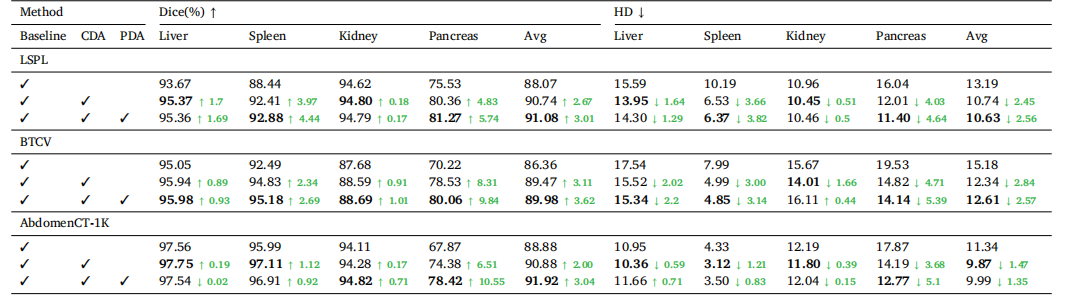
Table 4Ablation study of key components on LSPL dataset. CDA denotes cross-set data augmentation, and PDA denotes prototype-based distribution alignment. Green numbers indicatethe performance improvement over the baseline.
表4在 LSPL 数据集上关键组件的消融研究。CDA 表示跨集数据增强,PDA 表示基于原型的分布对齐。绿色数字表示相对于基线的性能提升。
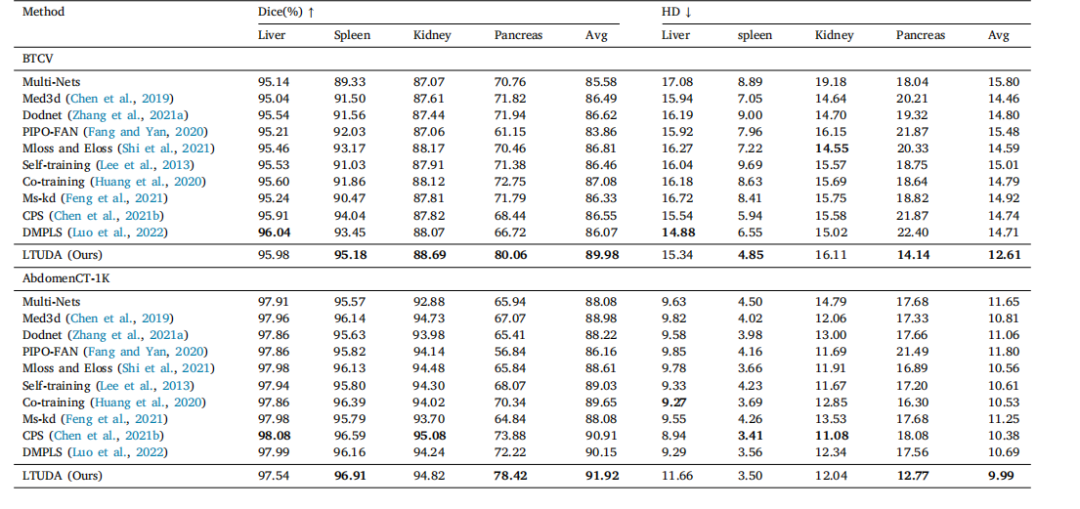
Table 5Generalization performance of different methods on two multi-organ datasets.
表5 在两个多脏器数据集上不同方法的泛化性能。
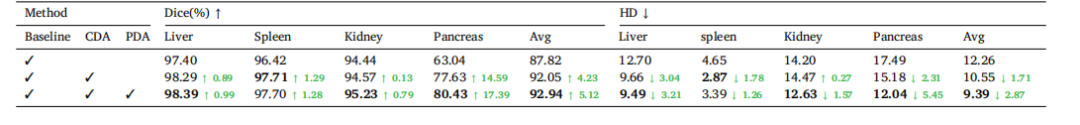
Table 6Ablation study of key components on the toy dataset. CDA denotes cross-set data augmentation, and PDA denotes prototype-based distribution alignment. Green numbers indicatethe performance improvement over the baseline.
表 6 在玩具数据集上关键组件的消融研究。CDA 表示跨集合数据增强,PDA 表示基于原型的分布对齐。绿色数字表示相对于基线的性能提升。
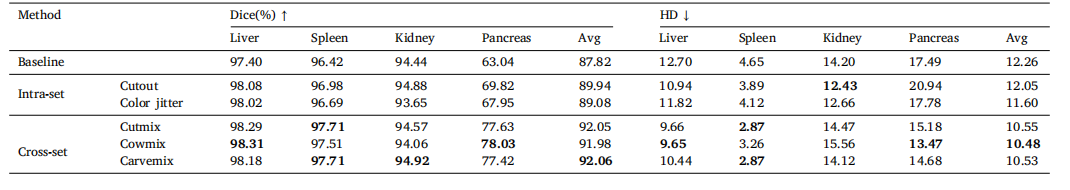
Table 7Ablation results of different data augmentation methods.
表 7 不同数据增强方法的消融结果。
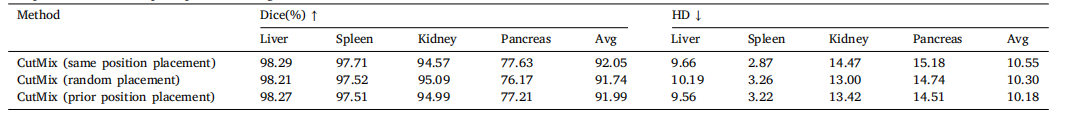
Table 8Comparison of different paste position strategies for CutMix
表 8 CutMix 不同粘贴位置策略的比较。
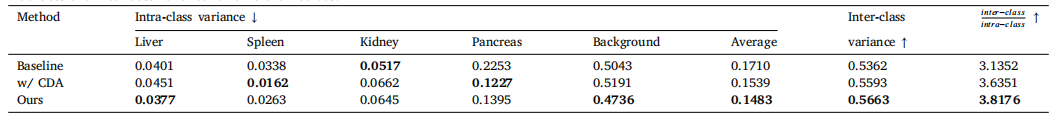
Table 9Intra-class and inter-class variance for different methods
表 9 不同方法的类内方差和类间方差。



















