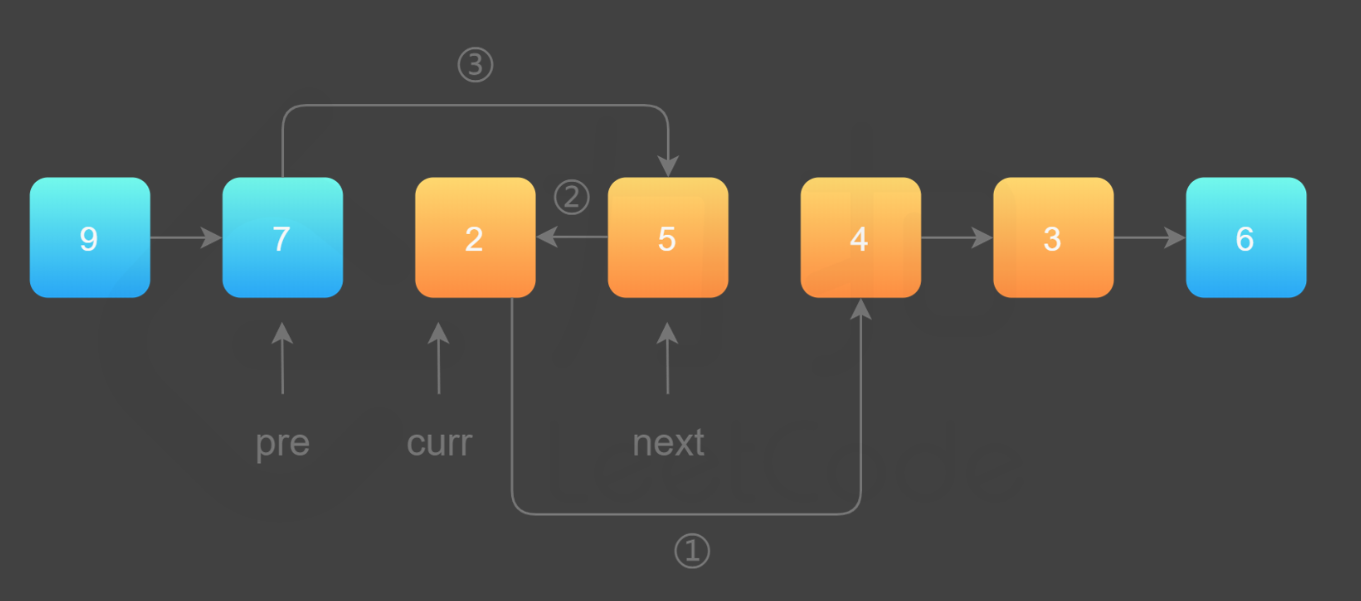
GATK(Genome Analysis Toolkit)是一个广泛使用的基因组分析工具包,它的核心库之一是htsjdk,用于处理高通量测序数据。在GATK中,ReadsPathDataSource类是负责管理和提供读取高通量测序数据文件(如BAM、SAM、CRAM)的类。
常见使用场景
- 数据加载:在GATK的基因组分析工具链中,
ReadsPathDataSource经常被用来从指定路径加载测序数据。 - 数据过滤:通过
ReadsPathDataSource,可以方便地在加载数据的同时进行预过滤,如按特定标准选择感兴趣的序列记录。 - 多文件支持:支持同时从多个文件中加载数据,使得分析多个样本的数据更加便捷。
类关系

ReadsPathDataSource源码
package org.broadinstitute.hellbender.engine;
import com.google.common.annotations.VisibleForTesting;
import htsjdk.samtools.MergingSamRecordIterator;
import htsjdk.samtools.SAMException;
import htsjdk.samtools.SAMFileHeader;
import htsjdk.samtools.SAMRecord;
import htsjdk.samtools.SAMSequenceDictionary;
import htsjdk.samtools.SamFileHeaderMerger;
import htsjdk.samtools.SamInputResource;
import htsjdk.samtools.SamReader;
import htsjdk.samtools.SamReaderFactory;
import htsjdk.samtools.util.CloseableIterator;
import htsjdk.samtools.util.IOUtil;
import org.apache.logging.log4j.LogManager;
import org.apache.logging.log4j.Logger;
import org.broadinstitute.hellbender.exceptions.GATKException;
import org.broadinstitute.hellbender.exceptions.UserException;
import org.broadinstitute.hellbender.utils.IntervalUtils;
import org.broadinstitute.hellbender.utils.SimpleInterval;
import org.broadinstitute.hellbender.utils.Utils;
import org.broadinstitute.hellbender.utils.gcs.BucketUtils;
import org.broadinstitute.hellbender.utils.iterators.SAMRecordToReadIterator;
import org.broadinstitute.hellbender.utils.iterators.SamReaderQueryingIterator;
import org.broadinstitute.hellbender.utils.read.GATKRead;
import org.broadinstitute.hellbender.utils.read.ReadConstants;
import java.io.IOException;
import java.nio.channels.SeekableByteChannel;
import java.nio.file.Path;
import java.util.ArrayList;
import java.util.Arrays;
import java.util.Iterator;
import java.util.LinkedHashMap;
import java.util.List;
import java.util.Map;
import java.util.Set;
import java.util.function.Function;
import java.util.stream.Collectors;
/**
* Manages traversals and queries over sources of reads which are accessible via {@link Path}s
* (for now, SAM/BAM/CRAM files only).
*
* Two basic operations are available:
*
* -Iteration over all reads, optionally restricted to reads that overlap a set of intervals
* -Targeted queries by one interval at a time
*/
public final class ReadsPathDataSource implements ReadsDataSource {
private static final Logger logger = LogManager.getLogger(ReadsPathDataSource.class);
/**
* Mapping from SamReaders to iterators over the reads from each reader. Only one
* iterator can be open from a given reader at a time (this is a restriction
* in htsjdk). Iterator is set to null for a reader if no iteration is currently
* active on that reader.
*/
private final Map<SamReader, CloseableIterator<SAMRecord>> readers;
/**
* Hang onto the input files so that we can print useful errors about them
*/
private final Map<SamReader, Path> backingPaths;
/**
* Only reads that overlap these intervals (and unmapped reads, if {@link #traverseUnmapped} is set) will be returned
* during a full iteration. Null if iteration is unbounded.
*
* Individual queries are unaffected by these intervals -- only traversals initiated via {@link #iterator} are affected.
*/
private List<SimpleInterval> intervalsForTraversal;
/**
* If true, restrict traversals to unmapped reads (and reads overlapping any {@link #intervalsForTraversal}, if set).
* False if iteration is unbounded or bounded only by our {@link #intervalsForTraversal}.
*
* Note that this setting covers only unmapped reads that have no position -- unmapped reads that are assigned the
* position of their mates will be returned by queries overlapping that position.
*
* Individual queries are unaffected by this setting -- only traversals initiated via {@link #iterator} are affected.
*/
private boolean traverseUnmapped;
/**
* Used to create a merged Sam header when we're dealing with multiple readers. Null if we only have a single reader.
*/
private final SamFileHeaderMerger headerMerger;
/**
* Are indices available for all files?
*/
private boolean indicesAvailable;
/**
* Has it been closed already.
*/
private boolean isClosed;
/**
* Initialize this data source with a single SAM/BAM file and validation stringency SILENT.
*
* @param samFile SAM/BAM file, not null.
*/
public ReadsPathDataSource( final Path samFile ) {
this(samFile != null ? Arrays.asList(samFile) : null, (SamReaderFactory)null);
}
/**
* Initialize this data source with multiple SAM/BAM files and validation stringency SILENT.
*
* @param samFiles SAM/BAM files, not null.
*/
public ReadsPathDataSource( final List<Path> samFiles ) {
this(samFiles, (SamReaderFactory)null);
}
/**
* Initialize this data source with a single SAM/BAM file and a custom SamReaderFactory
*
* @param samPath path to SAM/BAM file, not null.
* @param customSamReaderFactory SamReaderFactory to use, if null a default factory with no reference and validation
* stringency SILENT is used.
*/
public ReadsPathDataSource( final Path samPath, SamReaderFactory customSamReaderFactory ) {
this(samPath != null ? Arrays.asList(samPath) : null, customSamReaderFactory);
}
/**
* Initialize this data source with multiple SAM/BAM files and a custom SamReaderFac