文章目录
- week50 CGNN
- 摘要
- Abstract
- 0. 概述
- 1. 题目
- 2. Abstract
- 3. 网络结构
- 3.1 状态更新网络
- 3.2 method
- 4. 文献解读
- 4.1 Introduction
- 4.2 创新点
- 4.3 实验过程
- 5. 结论
- 6.相关代码
- CompositeLGNN
- CompositeGNN
- composite-graph-class
- 小结
- 参考文献
week50 CGNN
摘要
本周阅读了题为Composite Graph Neural Networks for Molecular Property Prediction的论文。该文探讨了复合图神经网络(cgnn)在分子分析任务中的能力。该文提出对原始循环GNN模型的标准版本和复合版本进行比较。该文使用了一些非常知名且可靠的基准测试数据集,它们是开放图基准测试(OGB)的一部分。每个数据集都由分子图组成。任务也由OGB定义,是基于分子活动或类别的分子分类和分子性质回归的混合。最后,实证证明了CGNN的有效性。
Abstract
This week’s weekly newspaper decodes the paper entitled Composite Graph Neural Networks for Molecular Property Prediction. This paper explores the capabilities of composite graph neural networks (CGNNs) in molecular analysis tasks. It proposes a comparison between the standard version and the composite version of the original recurrent GNN model. The study employs several highly reputable and reliable benchmark datasets that are part of the Open Graph Benchmark (OGB). Each dataset consists of molecular graphs. The tasks, also defined by OGB, are a mix of molecular classification based on molecular activities or categories and molecular property regression. Finally, the empirical results demonstrate the effectiveness of CGNNs.
0. 概述
- 分类和回归任务,创新点:复合图神经网络使用多个状态更新网络处理异构图,每个网络专用于特定节点类型
- 该文章具有回归任务,提供了使用基于异构图的GNN进行数值预测的理论与实践证明
1. 题目
标题:Composite Graph Neural Networks for Molecular Property Prediction.
作者:Bongini, P.; Pancino, N.; Bendjeddou, A.; Scarselli, F.; Maggini, M.; Bianchini, M.
发布:Int. J. Mol. Sci. 2024, 25, 6583.(中科院二区)
链接:https://doi.org/10.3390/ijms25126583
2. Abstract
分子是由不同种类的原子组成的异质图。复合图神经网络使用多个状态更新网络处理异构图,每个状态更新网络专用于特定的节点类型。这种方法可以比标准图神经网络更有效地从s图中提取信息,标准图神经网络通过一个单热编码类型的向量来区分节点类型。在8个分子图数据集上进行了广泛的实验,并进行了大量的分类和回归任务。实验得到的结果清楚地表明,在这种情况下,复合图神经网络的效率远远高于标准图神经网络。
3. 网络结构
gnn通过计算每个节点的状态来处理结构信息。节点状态应该获取节点本身的所有相关信息、局部图结构以及边缘和邻近节点的特征。在称为“消息传递”的迭代过程中,每个节点将其状态发送给所有邻居,并从所有邻居那里接收状态。为了计算下一个时刻(迭代)的状态值,所有传入的消息被聚合,将信息封装在它们所经过的边缘上。前一次迭代中的节点状态和聚合的消息作为输入馈送到状态更新网络,该网络在输出中产生新的节点状态。
3.1 状态更新网络
状态更新网络是一种神经网络(通常是MLP)。这个网络被复制到每个图节点上,作为一种构建块来组成GNN的体系结构(它回溯输入图结构)。状态更新网络的所有副本共享相同的权值,限制了模型参数的数量,防止了梯度消失问题。消息传递过程如图1所示。
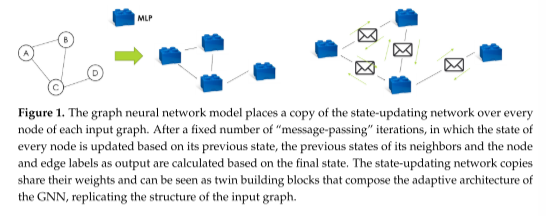
经过固定次数的消息传递迭代后,计算输出函数。为了做到这一点,在节点或边缘上复制专用的输出网络,类似于使用状态更新网络所执行的操作。输出网络是另一个MLP,它的所有副本共享相同的权重。如果问题以节点为中心,则输出网络将在每个节点(或图节点的相关子集)上复制。每个副本只接受相应节点的最终状态(在最后一次消息传递迭代之后)。如果问题以图为中心,则以相同的方式计算输出,然后通过对单个节点输出求和或平均,将输出聚合到整个图上。如果问题以边为中心,则输出网络在每个边(或图边的相关子集)上复制。在这种情况下,它的输入是由对应的边缘连接到边缘特征向量的两个节点的状态组成的。由所有状态更新MLP副本和输出MLP副本组成的网络称为编码网络;这在其体系结构中复制了输入图的结构。该网络也随时间展开,在每次消息传递迭代中复制每个状态更新MLP副本一次。后一步得到的结构称为未展开编码网络,它是通过结构算法进行反向传播的结构。
3.2 method
问题定义
给定一个图,G = (N, E),其中N是节点的集合, E = { ( n , m ) : n , m ∈ V } E = \{(n, m): n, m∈V\} E={(n,m):n,m∈V}表示边的集合,可以定义一个邻域函数 N e ( N ) = m ∈ N : ( n , m ) ∈ E N_e(N) = m∈N: (n, m)∈E Ne(N)=m∈N:(n,m)∈E,将每个节点映射到它的邻居的集合。节点可以与标签 l n ∀ n ∈ n l_n∀n∈n ln∀n∈n相关联,以描述它们的属性。边可以与标签 e m , n ∀ ( m , n ) ∈ E e_{m,n}∀(m, n)∈E em,n∀(m,n)∈E相关联,以描述相应的关系。
状态更新函数
定义一个GNN作为模型,近似一个输出函数,
g
w
g_w
gw可以定义在节点或其中的一个子集,
N
o
u
t
⊆
N
N_{out}⊆N
Nout⊆N,在这些边或者它们的子集上
E
o
u
t
⊆
E
E_{out}⊆E
Eout⊆E或者整个图上的G。在该文中,
g
w
g_w
gw总是被定义为整个图的一个性质,G。为了计算这个近似,GNN将处理图的结构以及节点和边的标签(特征)。GNN将一个状态
x
n
x_n
xn关联到每个节点,
n
∈
N
n∈N
n∈N,它将由一个可学习的状态更新函数fw迭代更新。状态是一个维数为
d
x
d_x
dx的向量,它被设置为GNN的一个超参数,通过从随机分布(通常以
R
d
x
\mathbb R^{d_x}
Rdx的原点为中心)中采样进行初始化。状态在K次迭代中更新,其中K是一个模型超参数。给定随机抽样的初始状态
x
n
0
,
∀
n
∈
N
x^0_n,∀n∈N
xn0,∀n∈N,则任意节点n在迭代t时的状态可以使用状态更新函数
f
w
f_w
fw来计算,如式(1)所示:
x
n
t
=
f
w
(
x
n
t
−
1
,
l
n
,
∑
m
∈
N
e
(
n
)
(
x
m
t
−
1
,
l
m
,
e
m
,
n
)
)
(1)
x^t_n=f_w(x^{t-1}_n,l_n,\sum_{m\in N_e(n)}(x^{t-1}_m,l_m,e_{m,n}))\tag{1}
xnt=fw(xnt−1,ln,m∈Ne(n)∑(xmt−1,lm,em,n))(1)
输出函数
当达到最大迭代次数K时,节点状态的最终版本
x
n
K
,
∀
n
∈
N
x^K_n,∀n∈N
xnK,∀n∈N,将作为输入输入到输出网络中,该网络近似于输出函数
g
w
g_w
gw。输出函数的形式取决于问题的类型。在本文中,只讨论以图为中心的问题,如式(2)所示:
y
G
=
1
∣
N
o
u
t
∣
∑
n
∈
N
o
u
t
g
w
(
x
n
K
,
l
n
)
(2)
y_G=\frac{1}{|N_{out}|}\sum_{n\in N_{out}}g_w(x^K_n,l_n) \tag{2}
yG=∣Nout∣1n∈Nout∑gw(xnK,ln)(2)
将状态更新函数迭代
然后在整个图中对所有节点的贡献取平均值,得到全局图输出
y
G
y_G
yG。这允许从多个不同的局部角度分析图形结构。然后将局部结果合并,获得对图的全局理解,包括图的结构以及节点和边的特征。在复合图神经网络(cgnn)中,存在多个状态更新网络,每个网络计算类型i的节点的状态更新函数的专用版本
f
w
,
i
f_{w,i}
fw,i。为了将每个节点映射到其类型,定义了一个函数T(n),将类型i关联到n。值得注意的是,所有状态更新函数fw,i具有相同的输出维数,对应于状态维数
d
x
d_x
dx;这允许不同类型的节点以无缝的方式交换消息。因此,状态更新函数可以改写为式(3):
x
n
T
=
f
w
,
i
(
x
n
t
−
1
,
l
n
,
a
∑
m
∈
N
e
(
n
)
(
x
m
t
−
1
,
e
m
,
n
)
)
:
i
=
T
(
n
)
(3)
x^T_n=f_{w,i}(x^{t-1}_n,l_n,a\sum_{m\in N_e(n)}(x^{t-1}_m,e_{m,n})):i=T(n)\tag{3}
xnT=fw,i(xnt−1,ln,am∈Ne(n)∑(xmt−1,em,n)):i=T(n)(3)
4. 文献解读
4.1 Introduction
该文将探讨复合图神经网络(cgnn)在分子分析任务中的能力。cgnn是原始递归GNN模型的一种变体,在递归GNN模型中,为图中每一种类型的节点定义一个状态更新网络。虽然gnn的预测能力已经得到了广泛的研究,但cgnn的预测能力目前还没有得到深入的分析。因此,该文提出对原始循环GNN模型的标准版本和复合版本进行比较。该文使用了一些非常知名且可靠的基准测试数据集,它们是开放图基准测试(OGB)的一部分。每个数据集都由分子图组成。任务也由OGB定义,是基于分子活动或类别的分子分类和分子性质回归的混合。在某些情况下,需要并行执行多个任务。
4.2 创新点
该文的主要贡献如下:
- 一种基于GNNkeras的综合方法,用于在任何分子图数据集上训练和评估cgnn和gnn的以图为中心的回归或分类任务;
- 在OGB的基准数据集上对cgnn和gnn进行了广泛的比较。
4.3 实验过程
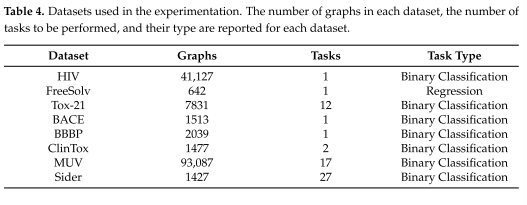
Dataset:OGB为每个基于图的任务提供不同大小的数据集。提供了分类和回归任务——关注节点、边和整个图。多类分类、多任务回归和链接预测问题也可用。此外,虽然通过OGB平台下载和评估,但所有数据集都来自MoleculeNet项目。分类和回归任务都得到了解决,有时在同一数据集上并行执行多个任务。表4列出了我们实验中使用的所有数据集及其主要特征,以及任务的数量和类型。
参数设置:
训练-验证-测试分割是基准测试提供的标准分割,并且在所有实验中都是相同的。此外,对于每个数据集,训练集示例、验证集示例和测试集示例的百分比分别为80%、10%和10%。earlystop-moudel的patience为10,在每个训练epoch之后应用。对OGB图进行预处理,将其作为gnn和cgnn的输入,分别转换为“GraphObjects”和“CompositeGraphObjects”。后两种数据类型由GNNkeras定义,并允许使用GNN和CGNN模型对图进行最佳处理。用表示原子种类的单热编码向量作为节点标签。对cgnn的节点类型和相应的单热编码进行了分组;这一步是必要的,以避免使用太多的节点类型和过长的单热向量。
评估标准:
使用OGB包提供的“评估器”进行评估;度量标准由OGB本身决定,并根据数据集而变化。
表1总结了在每个数据集上获得的最佳配置,报告了最佳模型的超参数。
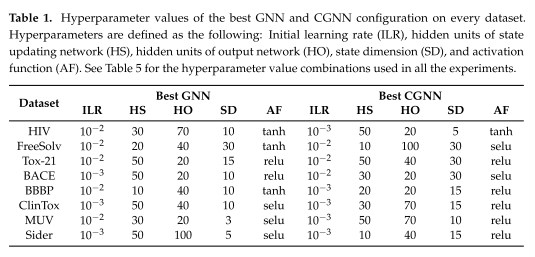
OGB Evaluator使用的评估指标是接收算子特征曲线下面积(AUROC)和平均精度(AP),用于二元分类问题,它使用均方根误差(RMSE)用于回归问题。
表2为最佳GNN和最佳CGNN配置的比较
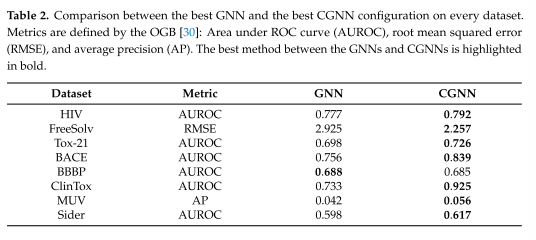
实验结果:
结果清楚地显示了CGNN在绝大多数分子数据集上的优势,BBBP是唯一的例外,只有0.003 AUROC差距。这证实了cgnn能够利用分子图的异质性,因此可以根据分子的活性更好地分类分子,并以较小的误差预测它们的性质。
由于状态更新网络在更精确的子任务上的专门化,使得cgnn得到了更好的结果。每个MLP专门化一种节点类型,允许在给定所有输入量(当前节点状态、邻居的当前状态、节点的标签、邻居和相对边)的情况下更准确地估计接下来的节点状态。mlp的这种专门化允许更好地局部理解图结构属性和节点/边缘标签,从而转化为输出网络更好的分类/回归性能(相反,它不专门化于节点类型)。这种好处带来了计算开销,由于使用了更多的mlp,模型的复杂性(就时间而言)增加了。
它仍然是用单个(非专业的)输出网络计算的。拥有一组输出网络,每个网络都专门在节点或边的类型上近似输出函数gw,预计将带来与不同状态更新网络所引入的优势成正比的优势
将这些结果扩展到以节点为中心和以边缘为中心的问题将是未来研究的问题。
表3中用于分析的任务的SotA方法,并将它们与CGNN模型进行了比较。
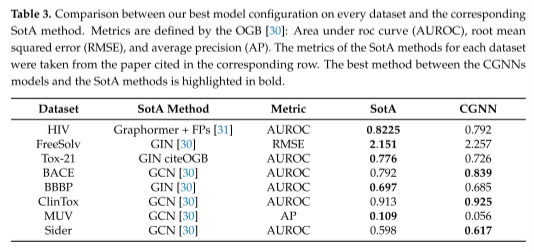
GINs[3]和GCNs[8]代表了大多数任务的最佳方法。相反,在HIV上,graphhormer的域特异性适应[31]优于所有其他方法。cgnn在8个数据集中的3个数据集上优于以前的SotA方法。特别是,它们在BACE、ClinTox和Sider上的表现优于GCNs。
在后一个数据集上,标准gnn与gcn完全相当,而cgnn的表现优于两者。这些实验结果符合预期,因为cgnn具有与标准gnn相同的理论特性,包括WL-1同构识别和图上的通用逼近,同时由于状态更新mlp的专业化,它们还利用了分子图的异构性质。这使得cgnn在大多数情况下能够优于WL-0 gcn,而后者对于某些类别的以图为中心的任务仍然更好,这要归功于循环GNNS和cgnn不可用的图池操作。
5. 结论
这项工作提出了标准循环GNN模型与CGNN模型之间的比较,前者处理异构图,并将不同物种的原子映射到不同的节点类型。实验证明,CGNN范式与循环gnn的近似能力相结合可以在各种场景中增强当前分子图的学习方法。这项工作的关键结论是,使用cgnn可以提高gnn在许多生物学问题上的能力。这带来了绝对可控的计算开销,因为与许多其他深度学习模型相比,gnn非常轻量,并且由于cgnn中包含的状态更新网络的专业化。
6.相关代码
https://doi.org/10.1016/j.softx.2022.101061
Pancino, N.; Bongini, P.; Scarselli, F.; Bianchini, M. GNNkeras: A Keras–based library for Graph Neural Networks and homogeneous and heterogeneous graph processing. SoftwareX 2022, 18, 101061. [CrossRef]
| v2.0 | |
|---|---|
| Permanent link to code/repository used for this code version | https://github.com/ElsevierSoftwareX/SOFTX-D-22-00019 |
CompositeLGNN
# codinf=utf-8
import tensorflow as tf
from GNN.Models.LGNN import LGNN
from GNN.Models.CompositeGNN import CompositeGNNnodeBased as GNNnodeBased, \
CompositeGNNarcBased as GNNarcBased, CompositeGNNgraphBased as GNNgraphBased
#######################################################################################################################
### CLASS LGNN - GENERAL ##############################################################################################
#######################################################################################################################
class CompositeLGNN(LGNN):
""" Composite Layered Graph Neural Network (CLGNN) model for node-focused, arc-focused or graph-focused applications. """
## REPR METHODs ###################################################################################################
def __repr__(self):
return f"Composite{super().__repr__()}"
## STATIC METHODs #################################################################################################
process_inputs = staticmethod(GNNnodeBased.process_inputs)
__gnnClass__ = staticmethod(lambda x: {GNNnodeBased:"node", GNNarcBased: "arc", GNNgraphBased: "graph"}[x])
__gnnClassLoader__ = staticmethod(lambda x: {"node": GNNnodeBased, "arc": GNNarcBased, "graph": GNNgraphBased}[x])
## LOOP METHODs ###################################################################################################
def Loop(self, nodes, arcs, dim_node_label, type_mask, set_mask, output_mask, composite_adjacencies, adjacency, arcnode, nodegraph,
training: bool = False) -> tuple[list[tf.Tensor], tf.Tensor, list[tf.Tensor]]:
""" Process a single GraphTensor element, returning 3 lists of iterations, states and outputs. """
constant_inputs = [type_mask, set_mask, output_mask, composite_adjacencies, adjacency, arcnode, nodegraph]
# get processing function to retrieve state and output for the nodes of the graphs processed by the gnn layer.
# Fundamental for graph-focused problem, since the output is referred to the entire graph, rather than to the graph nodes.
# Since in CONSTRUCTOR GNNs type must be only one, type(self.gnns[0]) is exploited and processing function is the same overall GNNs layers.
processing_function = self.__gnnClassLoader__("arc").Loop if self.gnns[0].name == "arc" else self.__gnnClassLoader__("node").Loop
# deep copy of nodes and arcs at time t==0.
dtype = tf.keras.backend.floatx()
nodes_0, arcs_0 = tf.constant(nodes, dtype=dtype), tf.constant(arcs, dtype=dtype)
# forward pass.
K, states, outs = list(), list(), list()
for idx, gnn in enumerate(self.gnns[:-1]):
# process graph.
k, state, out = processing_function(gnn, nodes, arcs, dim_node_label, *constant_inputs, training=training)
# append new k, new states and new gnn output.
K.append(k)
states.append(state)
outs.append(tf.sparse.sparse_dense_matmul(nodegraph, out, adjoint_a=True) if isinstance(gnn, GNNgraphBased) else out)
# update graph with nodes' state and nodes/arcs' output of the current GNN layer, to feed next GNN layer.
nodes, arcs, dim_node_label = self.update_graph(nodes_0, arcs_0, dim_node_label, set_mask, output_mask, state, out)
# final GNN k, state and out values.
k, state, out = self.gnns[-1].Loop(nodes, arcs, dim_node_label, *constant_inputs, training=training)
# return 3 lists of Ks, states and gnn outputs, s.t. len == self.LAYERS.
return K + [k], states + [state], outs + [out]
CompositeGNN
# codinf=utf-8
import tensorflow as tf
#######################################################################################################################
### CLASS COMPOSITE GNN - NODE BASED ##################################################################################
#######################################################################################################################
class CompositeGNNnodeBased(tf.keras.Model):
""" Composite Graph Neural Network (CGNN) model for node-focused applications. """
name = "node"
## CONSTRUCTORS METHODS ###########################################################################################
def __init__(self,
net_state: list[tf.keras.models.Sequential],
net_output: tf.keras.models.Sequential,
state_vect_dim: int,
max_iteration: int,
state_threshold: float) -> None:
""" CONSTRUCTOR
:param net_state: (list of tf.keras.model.Sequential) 1 MLP for each node type for the state networks, initialized externally.
:param net_output: (tf.keras.model.Sequential) MLP for the output network, initialized externally.
:param state_vect_dim: (int)>=0, dimension for state vectors in GNN where states_t0 != node labels.
:param max_iteration: (int) max number of iteration for the unfolding procedure to reach convergence.
:param state_threshold: (float) threshold for specifying if convergence is reached or not. """
assert state_vect_dim >= 0
assert max_iteration > 0
assert state_threshold >= 0
super().__init__(name=self.name)
# GNN parameters.
self.net_state = net_state
self.net_output = net_output
self.state_vect_dim = int(state_vect_dim)
self.max_iteration = int(max_iteration)
self.state_threshold = state_threshold
self.average_st_grads = None
# -----------------------------------------------------------------------------------------------------------------
def copy(self, copy_weights: bool = True):
""" COPY METHOD
:param copy_weights: (bool) True: state and output weights are copied in new gnn, otherwise they are re-initialized.
:return: a Deep Copy of the Composite GNN instance. """
# get configuration dictionary
config = self.get_config()
# MLPs
config["net_state"] = [tf.keras.models.clone_model(i) for i in config["net_state"]]
config["net_output"] = tf.keras.models.clone_model(config["net_output"])
if copy_weights:
for i, j in zip(config["net_state"], self.net_state): i.set_weights(j.get_weights())
config["net_output"].set_weights(self.net_output.get_weights())
# return copy
return self.from_config(config)
## CONFIG METHODs #################################################################################################
def get_config(self):
""" Get configuration dictionary. To be used with from_config().
It is good practice providing this method to user. """
return {"net_state": self.net_state,
"net_output": self.net_output,
"state_vect_dim": self.state_vect_dim,
"max_iteration": self.max_iteration,
"state_threshold": self.state_threshold}
# -----------------------------------------------------------------------------------------------------------------
@classmethod
def from_config(cls, config, **kwargs):
return cls(**config)
## REPRESENTATION METHODs #########################################################################################
def __repr__(self):
""" Representation string for the instance of Composite GNN. """
return f"Composite{super().__repr__()}"
# -----------------------------------------------------------------------------------------------------------------
def __str__(self):
""" Representation string for the instance of Composite GNN, for print() purpose. """
return self.__repr__()
## SAVE AND LOAD METHODs ##########################################################################################
def save(self, path: str, *args, **kwargs):
""" Save model to folder <path>.
:param path: (str) path in which model is saved.
:param args: args argument of tf.keras.models.save_model function.
:param kwargs: kwargs argument of tf.keras.models.save_model function. """
# check path
if path[-1] != '/': path += '/'
# get configuration dictionary.
config = self.get_config()
# save net_states and net_output.
for i, elem in enumerate(config.pop("net_state")):
tf.keras.models.save_model(elem, f'{path}net_state_{i}/', *args, **kwargs)
tf.keras.models.save_model(config.pop("net_output"), f'{path}net_output/', *args, **kwargs)
# save configuration (without MLPs info) file in json format.
from json import dump
with open(f'{path}config.json', 'w') as json_file:
dump(config, json_file)
# -----------------------------------------------------------------------------------------------------------------
@classmethod
def load(cls, path: str, *args, **kwargs):
""" Load model from folder <path>.
:param path: (str) path from which model is loaded.
:param args: args argument of tf.keras.models.load_model function.
:param kwargs: kwargs argument of tf.keras.models.load_model function. """
# check path
if path[-1] != '/': path += '/'
# load configuration file
from json import loads
with open(f'{path}config.json', 'r') as read_file:
config = loads(read_file.read())
# load net_state and net_output
from os import listdir
net_state_dirs = [f'{path}{i}/' for i in listdir(path) if 'net_state' in i]
netS = [tf.keras.models.load_model(i, compile=False, *args, **kwargs) for i in net_state_dirs]
netO = tf.keras.models.load_model(f'{path}net_output/', compile=False, *args, **kwargs)
return cls(net_state=netS, net_output=netO, **config)
## SUMMARY METHOD #################################################################################################
def summary(self, *args, **kwargs):
""" Summary method, to have a graphical representation for the Composite GNN model. """
super().summary(*args, **kwargs)
for net in self.net_state + [self.net_output]:
print('\n\n')
net.summary(*args, **kwargs)
## COMPILE METHOD #################################################################################################
def compile(self, *args, average_st_grads=False, **kwargs):
""" Configures the model for learning.
:param args: args inherited from Model.compile method. See source for details.
:param average_st_grads: (bool) If True, net_state params are averaged wrt the number of iterations, summed otherwise.
:param kwargs: Arguments supported for backwards compatibility only. Inherited from Model.compile method. See source for details.
:raise: ValueError – In case of invalid arguments for `optimizer`, `loss` or `metrics`. """
# force eager execution, since graph-mode must be implemented.
kwargs['run_eagerly'] = True
super().compile(*args, **kwargs)
for net in self.net_state: net.compile(*args, **kwargs)
self.net_output.compile(*args, **kwargs)
self.average_st_grads = average_st_grads
## CALL METHODs ###################################################################################################
def call(self, inputs, training: bool = False, mask=None):
""" Call method, get the output of the model for an input graph.
Return only output if testing mode
:param inputs: (tuple) coming from a GraphSequencer.__getitem__ method, since GNN cannot digest graph as they are.
:param training: (bool) True/False for training or testing mode, respectively.
:param mask: inherited from Model.call method. Useless here. Inserted just to avoid warning messages.
:return: only output of the model if training == False, or a tuple of 3 elements describing, respectively:
the iteration number reached at the end of Loop method at time T, the nodes state at time T and the output of the model. """
inputs = self.process_inputs(inputs)
k, state, out = self.Loop(*inputs, training=training)
if training: return k, state, out
else: return out
# -----------------------------------------------------------------------------------------------------------------
@staticmethod
def process_inputs(inputs):
""" convert some inputs in SparseTensor (not handled by default) and squeeze masks for correct computation. """
# get a list from :param inputs: tuple, so as to set elements in list (since a tuple is not settable).
inputs = list(inputs)
# squeeze inputs: [2] dim node labels, [3] type mask, [4] set mask, [5] output mask,
# to make them 1-dimensional (length,).
inputs[2:6] = [tf.squeeze(k, axis=-1) for k in inputs[2:6]]
# initialize sparse tensors -> [6] composite adjacency, [7] adjacency, [8] arcnode, [9] nodegraph.
inputs[6] = [tf.SparseTensor(indices=i, values=tf.squeeze(v, axis=-1), dense_shape=tf.squeeze(s)) for i, v, s in inputs[6]]
inputs[7:] = [tf.SparseTensor(k[0], values=tf.squeeze(k[1], axis=-1), dense_shape=tf.squeeze(k[2])) for k in inputs[7:]]
return inputs
## LOOP METHODS ###################################################################################################
def condition(self, k, state, state_old, *args) -> tf.bool:
""" Boolean function condition for tf.while_loop correct processing graphs. """
# distance_vector is the Euclidean Distance: √ Σ(xi-yi)² between current state xi and past state yi.
outDistance = tf.sqrt(tf.reduce_sum(tf.square(tf.subtract(state, state_old)), axis=1))
# state_norm is the norm of state_old, defined by ||state_old|| = √ Σxi².
state_norm = tf.sqrt(tf.reduce_sum(tf.square(state_old), axis=1))
# boolean vector that stores the "convergence reached" flag for each node.
scaled_state_norm = tf.math.scalar_mul(self.state_threshold, state_norm)
# check whether global convergence and/or the maximum number of iterations have been reached.
checkDistanceVec = tf.greater(outDistance, scaled_state_norm)
# compute boolean.
c1 = tf.reduce_any(checkDistanceVec)
c2 = tf.less(k, self.max_iteration)
return tf.logical_and(c1, c2)
# -----------------------------------------------------------------------------------------------------------------
def convergence(self, k, state, state_old, nodes, dim_node_label, type_mask, adjacency, aggregated_component, training) -> tuple:
""" Compute new state for the graph's nodes. """
# aggregated_states is the aggregation of ONLY neighbors' states.
aggregated_states = tf.sparse.sparse_dense_matmul(adjacency, state, adjoint_a=True)
# concatenate the destination node 'old' states to the incoming message, to obtain the input to net_state.
state_new = list()
for d, m, net in zip(dim_node_label, type_mask, self.net_state):
inp_state_i = tf.concat([nodes[:, :d], state, aggregated_states, aggregated_component], axis=1)
inp_state_i = tf.boolean_mask(inp_state_i, m)
# compute new state and update step iteration counter.
state_new.append(net(inp_state_i, training=training))
# reorder state based on nodes' ordering.
state_new = [tf.scatter_nd(tf.where(m), s, (len(m), s.shape[1])) for m, s in zip(type_mask, state_new)]
state_new = tf.reduce_sum(state_new, axis=0)
return k + 1, state_new, state, nodes, dim_node_label, type_mask, adjacency, aggregated_component, training
# -----------------------------------------------------------------------------------------------------------------
def apply_filters(self, state_converged, nodes, adjacency, arcs_label, mask) -> tf.Tensor:
""" Takes only nodes' [states] or [states|labels] for those with output_mask==1 AND belonging to set. """
return tf.boolean_mask(state_converged, mask)
# -----------------------------------------------------------------------------------------------------------------
def Loop(self, nodes, arcs, dim_node_label, type_mask, set_mask, output_mask, composite_adjacencies, adjacency,
arcnode, nodegraph, training: bool = False) -> tuple[int, tf.Tensor, tf.Tensor]:
""" Process a single GraphObject/GraphTensor element g, returning iteration, states and output. """
# get tensorflow dtype.
dtype = tf.keras.backend.floatx()
# initialize states and iters for convergence loop,
# including aggregated neighbors' label and aggregated incoming arcs' label.
aggregated_nodes = [tf.sparse.sparse_dense_matmul(a, nodes[:, :d], adjoint_a=True) for a, d in zip(composite_adjacencies, dim_node_label)]
aggregated_arcs = tf.sparse.sparse_dense_matmul(arcnode, arcs[:, 2:], adjoint_a=True)
aggregated_component = tf.concat(aggregated_nodes + [aggregated_arcs], axis=1)
# new values for Loop.
k = tf.constant(0, dtype=dtype)
if self.state_vect_dim > 0: state = tf.random.normal((nodes.shape[0], self.state_vect_dim), stddev=0.1, dtype=dtype)
else: state = tf.constant(nodes, dtype=dtype)
state_old = tf.ones_like(state, dtype=dtype)
training = tf.constant(training, dtype=bool)
# loop until convergence is reached.
k, state, state_old, *_ = tf.while_loop(self.condition, self.convergence,
[k, state, state_old, nodes, dim_node_label, type_mask, adjacency, aggregated_component, training])
# out_st is the converged state for the filtered nodes, depending on g.set_mask.
mask = tf.logical_and(set_mask, output_mask)
input_to_net_output = self.apply_filters(state, nodes, adjacency, arcs[:, 2:], mask)
# compute the output of the gnn network.
out = self.net_output(input_to_net_output, training=training)
return k, state, out
## TRAIN METHODS ##################################################################################################
def train_step(self, data):
""" training step used for fitting models. """
# Retrieve data from GraphSequencer.
x, y, sample_weight = data
# Run forward pass.
with tf.GradientTape() as tape:
k, state, y_pred = self(x, training=True)
loss = self.compiled_loss(y, y_pred, sample_weight, regularization_losses=self.losses)
if self.loss and y is None:
raise TypeError('Target data is missing. Your model was compiled with `loss` '
'argument and so expects targets to be passed in `fit()`.')
# Run backwards pass.
wS, wO = [j for i in self.net_state for j in i.trainable_variables], self.net_output.trainable_variables
dwbS, dwbO = tape.gradient(loss, [wS, wO])
if self.average_st_grads: dwbS = [i / k for i in dwbS]
self.optimizer.apply_gradients(zip(dwbS + dwbO, wS + wO))
self.compiled_metrics.update_state(y, y_pred, sample_weight)
# Collect metrics to return.
return_metrics = {}
for metric in self.metrics:
result = metric.result()
if isinstance(result, dict): return_metrics.update(result)
else: return_metrics[metric.name] = result
return return_metrics
#######################################################################################################################
### CLASS COMPOSITE GNN - EDGE BASED ##################################################################################
#######################################################################################################################
class CompositeGNNarcBased(CompositeGNNnodeBased):
""" Composite Graph Neural Network (CGNN) model for arc-focused applications. """
name = "arc"
## LOOP METHODS ###################################################################################################
def apply_filters(self, state_converged, nodes, adjacency, arcs_label, mask) -> tf.Tensor:
""" Takes only nodes' [states] or [states|labels] for those with output_mask==1 AND belonging to set. """
# gather source nodes' and destination nodes' state.
states = tf.gather(state_converged, adjacency.indices)
states = tf.reshape(states, shape=(arcs_label.shape[0], 2 * state_converged.shape[1]))
states = tf.cast(states, tf.keras.backend.floatx())
# concatenate source and destination states (and labels) to arc labels.
arc_state = tf.concat([states, arcs_label], axis=1)
# takes only arcs states for those with output_mask==1 AND belonging to the set (in case Dataset == 1 Graph).
return tf.boolean_mask(arc_state, mask)
#######################################################################################################################
### CLASS COMPOSITE GNN - GRAPH BASED #################################################################################
#######################################################################################################################
class CompositeGNNgraphBased(CompositeGNNnodeBased):
""" Composite Graph Neural Network (CGNN) model for graph-focused applications. """
name = "graph"
## LOOP METHODS ###################################################################################################
def Loop(self, *args, training: bool = False) -> tuple[int, tf.Tensor, tf.Tensor]:
""" Process a single graph, returning iteration, states and output.
Output of graph-focused problem is the averaged nodes output. """
k, state_nodes, out_nodes = super().Loop(*args, training=training)
out_gnn = tf.sparse.sparse_dense_matmul(args[-1], out_nodes, adjoint_a=True)
return k, state_nodes, out_gnn
composite-graph-class
# coding=utf-8
import sys
import numpy as np
import tensorflow as tf
from scipy.sparse import coo_matrix
from GNN.graph_class import GraphObject, GraphTensor
#######################################################################################################################
## COMPOSITE GRAPH OBJECT CLASS #######################################################################################
#######################################################################################################################
class CompositeGraphObject(GraphObject):
""" Heterogeneous Graph data representation. Composite GNNs are based on this class. """
## CONSTRUCTORS METHODs ###########################################################################################
def __init__(self, nodes, arcs, targets, type_mask, dim_node_label, *args, **kwargs):
""" CONSTRUCTOR METHOD
:param nodes: Ordered Nodes Matrix X where nodes[i, :] = [i-th node Label].
:param arcs: Ordered Arcs Matrix E where arcs[i, :] = [From ID Node | To ID Node | i-th arc Label].
:param targets: Targets Matrix T with shape (Num of arcs/node targeted example or 1, dim_target example).
:param type_mask: boolean np.array with shape (Num of nodes, Num of node's types). type_mask[:,i] refers to dim_node_label[i].
:param dim_node_label: (list/tuple) with len == Num of node's types. i-th element defines label dimension of nodes of type i.
:param focus: (str) The problem on which graph is used: 'a' arcs-focused, 'g' graph-focused, 'n' node-focused.
:param set_mask: Array of boolean {0,1} to define arcs/nodes belonging to a set, when dataset == single GraphObject.
:param output_mask: Array of boolean {0,1} to define the sub-set of arcs/nodes whose target is known.
:param sample_weight: target sample weight for loss computation. It can be int, float or numpy.array of ints or floats:
> If int or float, all targets are weighted as sample_weight * ones.
> If numpy.array, len(sample_weight) and targets.shape[0] must agree.
:param ArcNode: Sparse matrix of shape (num_of_arcs, num_of_nodes) s.t. A[i,j]=value if arc[i,2]==node[j].
:param NodeGraph: Sparse matrix in coo format of shape (nodes.shape[0], {Num graphs or 1}) used only when focus=='g'.
:param aggregation_mode: (str) The aggregation mode for the incoming message based on ArcNode and Adjacency matrices:
---> elem(matrix)={0-1};
> 'average': A'X gives the average of incoming messages, s.t. sum(A[:,i])==1;
> 'normalized': A'X gives the normalized message wrt the total number of g.nodes, s.t. sum(A)==1;
> 'sum': A'X gives the total sum of incoming messages, s.t. A={0,1}.
> 'composite_average': A'X gives the average of incoming messages wrt node's type, s.t. sum(A[:,i])>=1. """
# type_mask[:,i] refers to nodes with DIM_NODE_LABEL[i] label dimension.
# Be careful when initializing a new graph!
self.type_mask = type_mask.astype(bool)
# AFTER initializing type_mask because of self.buildAdjacency method.
super().__init__(nodes, arcs, targets, *args, **kwargs)
# store dimensions: first two columns of arcs contain nodes indices.
self.DIM_NODE_LABEL = np.array(dim_node_label, ndmin=1, dtype=int)
# build Composite Adjacency Matrices. It is a list of Adjacency Matrix as long as the number of nodes' types.
# i-th element corresponds to a composite matrix where only nodes' type 'i' is considered.
# ADJ[k][i,j]=value if and only if an edge (i,j) exists AND node_type(i) == k.
self.CompositeAdjacencies = self.buildCompositeAdjacency()
# -----------------------------------------------------------------------------------------------------------------
def buildCompositeAdjacency(self):
""" Build a list ADJ of Composite Aggregated Adjacency Matrices,
s.t. ADJ[t][i,j]=value if an edge (i,j) exists AND type(i)==k.
:return: list of sparse Matrices in coo format, for memory efficiency. One for each node's type. """
composite_adjacencies = [self.Adjacency.copy() for _ in range(len(self.DIM_NODE_LABEL))]
# set to 0 rows of nodes of incorrect type.
for t, a in zip(self.type_mask.transpose(), composite_adjacencies):
not_type_node_mask = np.in1d(self.arcs[:, 0], np.argwhere(t), invert=True)
a.data[not_type_node_mask] = 0
a.eliminate_zeros()
return composite_adjacencies
# -----------------------------------------------------------------------------------------------------------------
def buildArcNode(self, aggregation_mode):
""" Build ArcNode Matrix A of shape (number_of_arcs, number_of_nodes) where A[i,j]=value if arc[i,2]==node[j].
Compute the matmul(m:=message,A) to get the incoming message on each node, composed of nodes' states and arcs' labels.
:return: sparse ArcNode Matrix in coo format, for memory efficiency.
:raise: Error if <aggregation_mode> is not in ['average', 'sum', 'normalized', 'composite_average']."""
if aggregation_mode not in ['normalized', 'average', 'sum', 'composite_average']: raise ValueError("ERROR: Unknown aggregation mode")
# initialize matrix. It's useless, just for not having any warning message at the end of the method.
matrix = None
# exploit super function.
if aggregation_mode in ['normalized', 'average', 'sum']:
matrix = super().buildArcNode(aggregation_mode)
# composite average node aggregation - incoming message as sum of averaged type-focused neighbors state,
# e.g. if a node i has 3 neighbors (2 of them belonging to a type k1, the other to a type k2):
# the message coming from k1's nodes is divided by 2,
# while the message coming from k2's node is taken as is, being that the only one neighbor belonging to k2.
elif aggregation_mode == 'composite_average':
# sum node aggregation - incoming message as sum of neighbors states and labels, then process composite average.
matrix = super().buildArcNode('sum')
# set to 0 rows of nodes of incorrect type.
for t in self.type_mask.transpose():
if not np.any(t): continue
type_node_mask = np.in1d(self.arcs[:, 0], np.argwhere(t), invert=False)
val, col_index, destination_node_counts = np.unique(matrix.col[type_node_mask], return_inverse=True, return_counts=True)
matrix.data[type_node_mask] /= destination_node_counts[col_index]
return matrix
# -----------------------------------------------------------------------------------------------------------------
def copy(self):
""" COPY METHOD
:return: a Deep Copy of the GraphObject instance. """
return CompositeGraphObject(arcs=self.getArcs(), nodes=self.getNodes(), targets=self.getTargets(),
set_mask=self.getSetMask(), output_mask=self.getOutputMask(),
sample_weight=self.getSampleWeights(), NodeGraph=self.getNodeGraph(),
aggregation_mode=self.aggregation_mode, dim_node_label=self.DIM_NODE_LABEL,
type_mask=self.getTypeMask())
## REPRESENTATION METHODs #########################################################################################
def __repr__(self):
""" Representation string of the instance of CompositeGraphObject. """
return f"composite_{super().__repr__()}"
## SETTERS ########################################################################################################
def setAggregation(self, aggregation_mode: str):
""" Set ArcNode values for the specified :param aggregation_mode: """
super().setAggregation(aggregation_mode)
self.CompositeAdjacencies = self.buildCompositeAdjacency()
## GETTERS ########################################################################################################
# ALL return a deep copy of the corresponding element.
def getTypeMask(self):
return self.type_mask.copy()
## SAVER METHODs ##################################################################################################
def get_dict_data(self):
""" Return all useful elements for storing a graph :param g:, in a dict format. """
data = super().get_dict_data()
data['type_mask'] = self.type_mask
data['dim_node_label'] = self.DIM_NODE_LABEL
return data
## CLASS METHODs ### MERGER #######################################################################################
@classmethod
def merge(cls, glist, focus: str, aggregation_mode: str, dtype='float32'):
""" Method to merge a list of CompositeGraphObject elements in a single GraphObject element.
:param glist: list of CompositeGraphObject elements to be merged.
> NOTE if focus=='g', new NodeGraph will have dimension (Num nodes, Num graphs).
:param aggregation_mode: (str) incoming message aggregation mode. See BuildArcNode for details.
:param dtype: dtype of elements of new arrays after merging procedure.
:return: a new CompositeGraphObject containing all the information (nodes, arcs, targets, ...) in glist. """
# get new GraphObject, then convert to CompositeGraphObject.
g = super().merge(glist, focus, 'sum', dtype)
dim_node_label, type_mask = zip(*[(i.DIM_NODE_LABEL, i.getTypeMask()) for i in glist])
# check if every graphs has the same DIM_NODE_LABEL attribute.
dim_node_label = set(tuple(i) for i in dim_node_label)
assert len(dim_node_label) == 1, "DIM_NODE_LABEL not unique among graphs in :param glist:"
# get single matrices for new graph.
type_mask = np.concatenate(type_mask, axis=0, dtype=bool)
# resulting CompositeGraphObject.
return CompositeGraphObject(arcs=g.arcs, nodes=g.nodes, targets=g.targets, type_mask=type_mask,
dim_node_label=dim_node_label.pop(), focus=focus,
set_mask=g.set_mask, output_mask=g.output_mask, sample_weight=g.sample_weight,
NodeGraph=g.NodeGraph, aggregation_mode=aggregation_mode)
## CLASS METHODs ### UTILS ########################################################################################
@classmethod
def fromGraphTensor(cls, g, focus: str):
""" Create CompositeGraphObject from CompositeGraphTensor.
:param g: a CompositeGraphTensor element to be translated into a CompositeGraphObject element.
:param focus: (str) 'n' node-focused; 'a' arc-focused; 'g' graph-focused. See __init__ for details.
:return: a CompositeGraphObject element whose tensor representation is g.
"""
nodegraph = coo_matrix((g.NodeGraph.values, tf.transpose(g.NodeGraph.indices))) if focus == 'g' else None
return cls(arcs=g.arcs.numpy(), nodes=g.nodes.numpy(), targets=g.targets.numpy(),
dim_node_label=g.DIM_NODE_LABEL.numpy(), type_mask=g.type_mask, set_mask=g.set_mask.numpy(),
output_mask=g.output_mask.numpy(), sample_weight=g.sample_weight.numpy(), NodeGraph=nodegraph,
aggregation_mode=g.aggregation_mode, focus=focus)
#######################################################################################################################
## COMPOSITE GRAPH TENSOR CLASS #######################################################################################
#######################################################################################################################
class CompositeGraphTensor(GraphTensor):
""" Tensor version of a CompositeGraphObject. Useful to speed up learning processes. """
## CONSTRUCTORS METHODs ###########################################################################################
def __init__(self, *args, type_mask, CompositeAdjacencies, **kwargs):
""" It contains all information to be passed to GNN model,
but described with tensorflow dense/sparse tensors. """
super().__init__(*args, **kwargs)
# constant tensors + sparse tensors.
self.type_mask = tf.constant(type_mask, dtype=bool)
self.CompositeAdjacencies = [tf.sparse.SparseTensor.from_value(i) for i in CompositeAdjacencies]
# -----------------------------------------------------------------------------------------------------------------
def copy(self):
""" COPY METHOD
:return: a Deep Copy of the CompositeGraphTensor instance. """
return CompositeGraphTensor(nodes=self.nodes, dim_node_label=self.DIM_NODE_LABEL, arcs=self.arcs,
targets=self.targets, set_mask=self.set_mask, output_mask=self.output_mask,
sample_weight=self.sample_weight, Adjacency=self.Adjacency, ArcNode=self.ArcNode,
NodeGraph=self.NodeGraph, aggregation_mode=self.aggregation_mode,
type_mask=self.type_mask, CompositeAdjacencies=self.CompositeAdjacencies)
## REPRESENTATION METHODs #########################################################################################
def __repr__(self):
""" Representation string for the instance of CompositeGraphTensor. """
return f"composite_{super().__repr__()}"
## STATIC METHODs ### SAVER #######################################################################################
@staticmethod
def save_graph(graph_path: str, g, compressed: bool = False, **kwargs) -> None:
""" Save a graph in a .npz compressed/uncompressed archive.
:param graph_npz_path: path where a single .npz file will be stored, for saving the graph.
:param g: graph of type GraphObject to be saved.
:param compressed: bool, if True graph will be stored in a compressed npz file, npz uncompressed otherwise.
:param kwargs: kwargs argument for for numpy.savez/numpy.savez_compressed function. """
data = {'type_mask': g.type_mask}
name = 'CompositeAdjacencies_'
for idx, mat in enumerate(g.CompositeAdjacencies):
data[f"{name}{idx}"] = tf.concat([mat.values[:, tf.newaxis], tf.cast(mat.indices, g.dtype)], axis=1)
super().save_graph(g, compressed, **kwargs, **data)
## CLASS METHODs ### LOADER #######################################################################################
@classmethod
def load(cls, graph_npz_path, **kwargs):
""" Load a GraphTensor from a npz compressed/uncompressed file.
:param graph_npz_path: path to the npz graph file.
:param kwargs: kwargs argument of numpy.load function. """
if '.npz' not in graph_npz_path: graph_npz_path += '.npz'
data = dict(np.load(graph_npz_path, **kwargs))
data['aggregation_mode'] = str(data['aggregation_mode'])
for i in ['Adjacency', 'ArcNode', 'NodeGraph']:
data[i] = tf.SparseTensor(indices=data[i][:,1:], values=data[i][:,0], dense_shape=data.pop(i + '_shape'))
CA = [data.pop(f"CompositeAdjacencies_{idx}") for idx, elem in enumerate(data['dim_node_label'])]
CA = [tf.SparseTensor(indices=adj[:,1:], values=adj[:,0], dense_shape=data['Adjacency'].shape) for adj in CA]
return cls(**data, CompositeAdjacencies=CA)
## CLASS and STATIC METHODs ### UTILS #############################################################################
@classmethod
def fromGraphObject(cls, g: CompositeGraphObject):
""" Create CompositeGraphTensor from CompositeGraphObject.
:param g: a CompositeGraphObject element to be translated into a CompositeGraphTensor element.
:return: a CompositeGraphTensor element whose normal representation is g. """
return cls(nodes=g.nodes, dim_node_label=g.DIM_NODE_LABEL, arcs=g.arcs, targets=g.targets, set_mask=g.set_mask,
output_mask=g.output_mask, sample_weight=g.sample_weight, Adjacency=cls.COO2SparseTensor(g.Adjacency),
ArcNode=cls.COO2SparseTensor(g.ArcNode), NodeGraph=cls.COO2SparseTensor(g.NodeGraph),
aggregation_mode=g.aggregation_mode, type_mask=g.type_mask.transpose(),
CompositeAdjacencies=[cls.COO2SparseTensor(i) for i in g.CompositeAdjacencies])
小结
这项工作提出了标准循环GNN模型与CGNN模型之间的比较,前者处理异构图,并将不同物种的原子映射到不同的节点类型。实验证明,CGNN范式与循环gnn的近似能力相结合可以在各种场景中增强当前分子图的学习方法。这项工作的关键结论是,使用cgnn可以提高gnn在许多生物学问题上的能力。这带来了绝对可控的计算开销,因为与许多其他深度学习模型相比,gnn非常轻量,并且由于cgnn中包含的状态更新网络的专业化。
参考文献
[1] Bongini, P.; Pancino, N.; Bendjeddou, A.; Scarselli, F.; Maggini, M.; Bianchini, M. Composite
Graph Neural Networks for Molecular Property Prediction. Int. J. Mol. Sci. 2024, 25, 6583. https://doi.org/10.3390/ijms25126583
[2] Pancino, N.; Bongini, P.; Scarselli, F.; Bianchini, M. GNNkeras: A Keras–based library for Graph Neural Networks and homogeneous and heterogeneous graph processing. SoftwareX 2022, 18, 101061. [CrossRef]