一位老师聊起火山图(Volcano plot | 别再问我这为什么是火山图 (在线轻松绘制)),说见过倾斜45度的类似图,可否演示怎么画?想了下,可能是下面这种图,绘起来看看。
检查和安装包
a = rownames(installed.packages())
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager", repos = site)
a = rownames(installed.packages())
install_bioc <-
c(
"egg",
"ggpubr",
"ggplot2"
)
for (i in install_bioc) {
if (!i %in% a)
BiocManager::install(i, update = F)
}
if (!"ImageGP" %in% a){
# devtools::install_github("Tong-Chen/ImageGP")
devtools::install_git("https://gitee.com/ct5869/ImageGP.git")
}
library(ImageGP)
library(ggplot2)
library(ggpubr)
library(egg)读入数据并标记差异基因
数据是基于DESeq2分析的差异基因数据。
file <- "data/ehbio.simplier.DESeq2.trt._vs_.untrt.results.txt"
diffexpr <- sp_readTable(file, row.names=1)
# 做一个log转换
diffexpr$trt <- log2(diffexpr$trt+1)
diffexpr$untrt <- log2(diffexpr$untrt+1)
head(diffexpr)
## trt untrt baseMean log2FoldChange pvalue padj
## ENSG00000152583 10.88130 6.354646 983.042 4.546 1.219e-91 2.149e-87
## ENSG00000189221 12.09256 8.706410 2391.559 3.387 9.955e-61 8.779e-57
## ENSG00000179094 10.40316 7.343044 757.249 3.065 2.435e-54 1.432e-50
## ENSG00000116584 10.50370 11.567470 2242.427 -1.064 3.957e-49 1.745e-45
## ENSG00000120129 12.55314 9.579082 3386.426 2.975 1.930e-48 6.807e-45
## ENSG00000134686 12.04738 10.587715 2884.847 1.460 1.846e-45 5.427e-42标记差异基因
diffexpr$level <- ifelse(diffexpr$padj<0.05,
ifelse(diffexpr$log2FoldChange>=1, "trt up",
ifelse(diffexpr$log2FoldChange<=-1, "untrt up", "NoSig")),"NoSig")
head(diffexpr)
## trt untrt baseMean log2FoldChange pvalue padj level
## ENSG00000152583 10.88130 6.354646 983.042 4.546 1.219e-91 2.149e-87 trt up
## ENSG00000189221 12.09256 8.706410 2391.559 3.387 9.955e-61 8.779e-57 trt up
## ENSG00000179094 10.40316 7.343044 757.249 3.065 2.435e-54 1.432e-50 trt up
## ENSG00000116584 10.50370 11.567470 2242.427 -1.064 3.957e-49 1.745e-45 untrt up
## ENSG00000120129 12.55314 9.579082 3386.426 2.975 1.930e-48 6.807e-45 trt up
## ENSG00000134686 12.04738 10.587715 2884.847 1.460 1.846e-45 5.427e-42 trt up基于ImageGP绘图
p <- sp_scatterplot(diffexpr, xvariable = "trt", yvariable = "untrt",
color_variable = "level",
color_variable_order = c("NoSig","trt up", "untrt up"),
manual_color_vector = c("grey","firebrick","dodgerblue"),
legend.position = c(0.8,0.3)) + coord_fixed(1)
p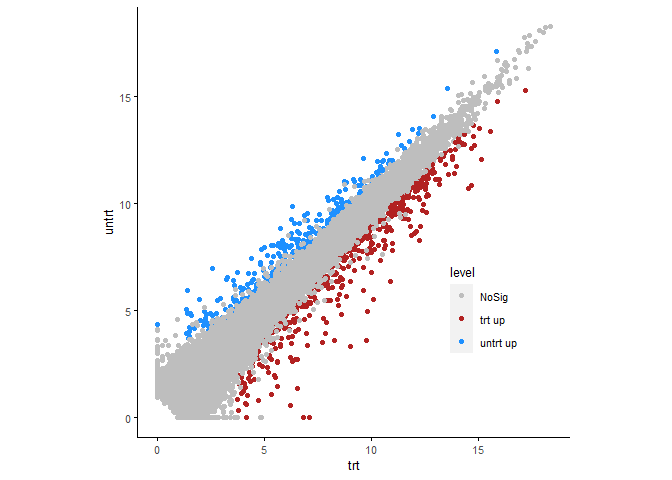
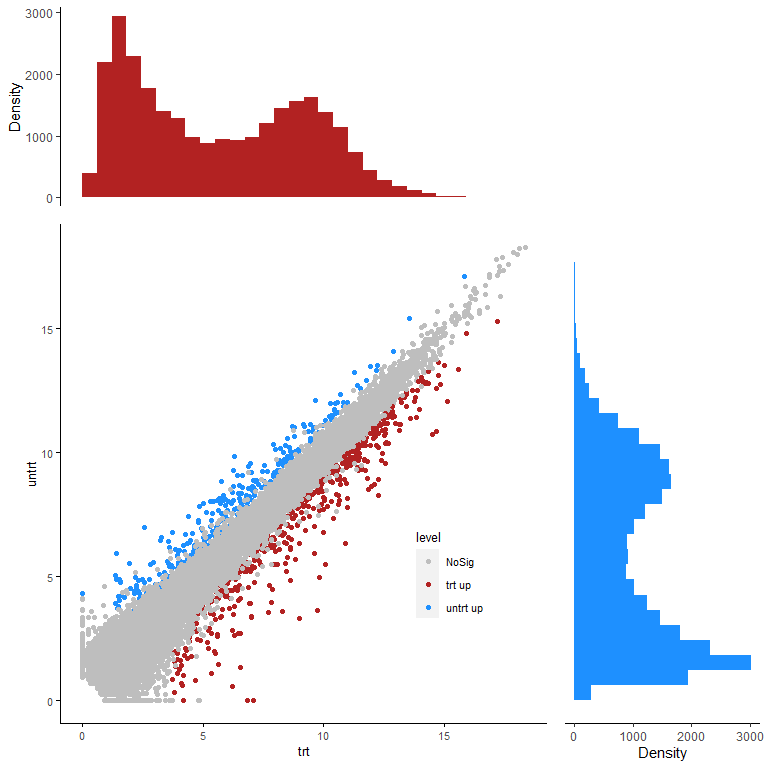
到这满足需求了,又有老师说能不能加上数据分布展示?
拼上Marginal plot
xplot <- ggplot(diffexpr, aes(x=trt)) + geom_histogram(fill="firebrick") +
theme_classic() +
theme(axis.line.x=element_blank(),
axis.ticks.x=element_blank(),
axis.text.x = element_blank(),
axis.title.x = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_blank(),
panel.background = element_blank(),
legend.title = element_blank(),
legend.position = c(0.75,0.85),
#legend.title = element_text(size = 5),
legend.text = element_text(size = 8),
legend.key.size = unit(0.5, "lines"),
legend.spacing = unit(0.3, "cm"),
) + ylab("Density")
xplot
yplot <- ggplot(diffexpr, aes(x=untrt)) + geom_histogram(fill="dodgerblue") +
theme_classic() +
theme(axis.line.y=element_blank(),
axis.ticks.y=element_blank(),
axis.text.y = element_blank(),
axis.title.y=element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_blank(),
panel.background = element_blank(),
legend.title = element_blank(),
legend.position = c(0.7,0.8),
#legend.title = element_text(size = 5),
legend.text = element_text(size = 8),
legend.key.size = unit(0.5, "lines"),
legend.spacing = unit(0.3, "cm"),
) + ylab("Density") +
rotate()
yplot
合并起来
white <- ggplot() + theme_void()
egg::ggarrange(xplot, white, p, yplot,
widths=c(5,2),heights = c(2,5),padding=unit(0,"line"))
也可以用现成的工具
用ggpubr绘制
ggscatterhist(diffexpr, x="trt", y="untrt", color="level",
palette=c("grey","firebrick","dodgerblue"), margin.plot="histogram" )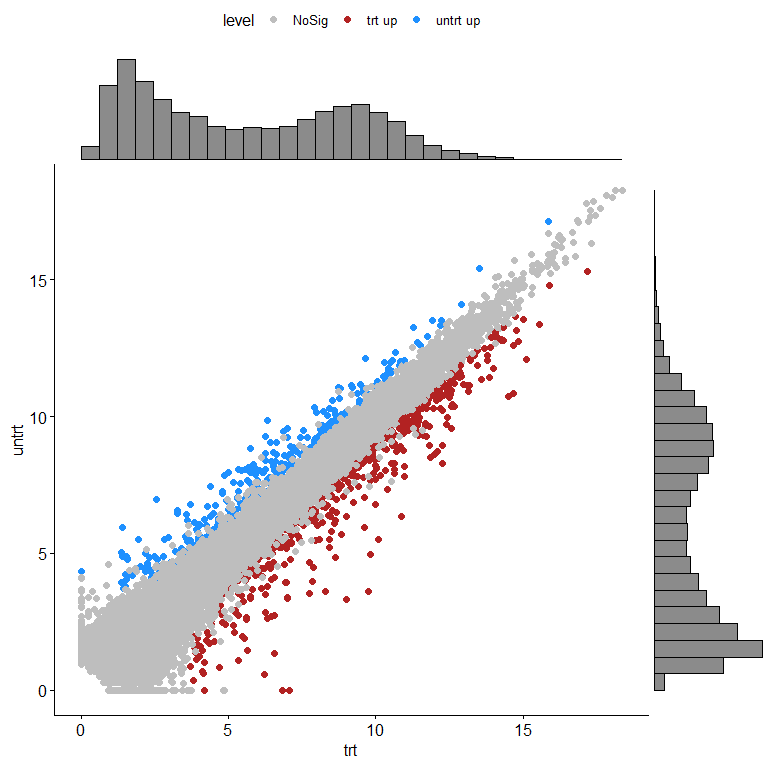
输出数据用在线工具绘制
见次条
高颜值免费在线绘图工具
http://www.ehbio.com/Cloud_Platform/front/#/analysis?page=b%27MTI%3D%27
sp_writeTable(diffexpr, file="DE_gene_label.txt")