简介
该宏基因组流程主要有四步,分别是1.检查raw data;2.获得高质量reads;3.合并PE数据;4. reads map到参考数据库得到profile。
初步想法:
- 先分别撰写每一步的基础脚本,如过滤,mapping等过程的脚本,只针对单样本;与此同时,设计好输入文件的格式;
- 接着脚本内部每个样本生成每个步骤的脚本,如sample1.trim.sh sample1.map.sh
- 然后将每步的脚本放置一起形成该步骤的综合脚本,如 step1.trim.sh
- 最后将含有每样本的各步骤的脚本综合在一起,为Run.all.sh
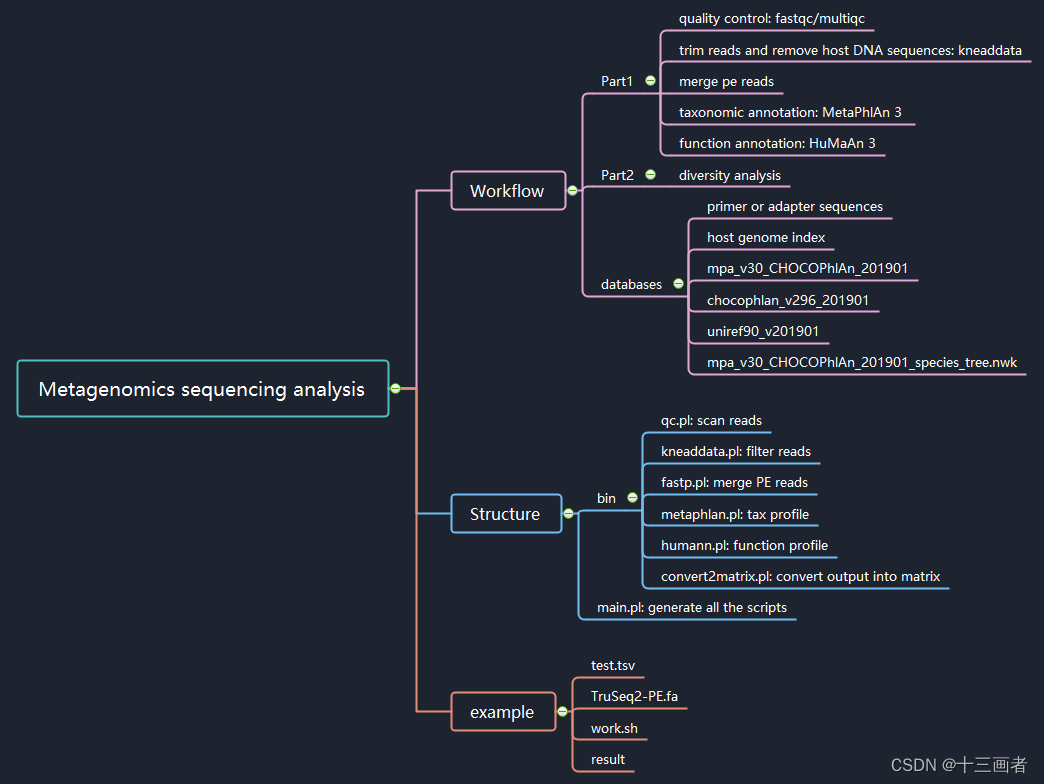
文件结构:脚本和结果文件
../MetaGenomics_pipeline/
├── bin # 脚本
│ ├── humann.pl
│ ├── kneaddata.pl
│ ├── merge.pl
│ ├── metaphlan.pl
│ └── qc.pl
├── main.pl # 主程序
├── result # 结果
│ ├── 00.quality
│ │ ├── fastqc
│ │ └── multiqc
│ ├── 01.kneaddata
│ ├── 02.merge
│ ├── 03.humann
│ │ ├── genefamilies
│ │ ├── log
│ │ ├── metaphlan
│ │ ├── pathabundance
│ │ └── pathcoverage
│ ├── 04.metaphlan
│ ├── Run.s1.qc.sh
│ ├── Run.s2.kneaddata.sh
│ ├── Run.s3.merge.sh
│ ├── Run.s4.humann.sh
│ ├── Run.s5.metaphlan.sh
│ └── script # 每个样本的每一步脚本
│ ├── 00.quality
│ ├── 01.kneaddata
│ ├── 02.merge
│ ├── 03.humann
│ └── 04.metaphlan
├── Run.all.sh
├── test.tsv
├── TruSeq2-PE.fa -> /data/share/anaconda3/share/trimmomatic/adapters/TruSeq2-PE.fa
└── work.sh # 启动脚本
步骤
先准备输入数据
find /RawData/ -name "*fq.gz" | sort | perl -e 'print "SampleID\tLaneID\tPath\n"; while(<>){chomp; $fq=(split("\/", $_))[-1]; $sampleid=$fq; $laneid=$fq; $sampleid=~s/\_R[1|2]\.fq.gz//g; $laneid=~s/\.fq.gz//g;print "$sampleid\t$laneid\t$_\n";}' > samples.fqpath.tsv
| SampleID | LaneID | Path |
|---|---|---|
| ND2 | ND2_R1 | RawData/ND2_R1.fq.gz |
| ND2 | ND2_R2 | RawData/ND2_R2.fq.gz |
| XL10 | XL10_R1 | RawData/XL10_R1.fq.gz |
| XL10 | XL10_R2 | RawData/XL10_R2.fq.gz |
| XL11 | XL11_R1 | RawData/XL11_R1.fq.gz |
| XL11 | XL11_R2 | RawData/XL11_R2.fq.gz |
| XL1 | XL1_R1 | RawData/XL1_R1.fq.gz |
| XL1 | XL1_R2 | RawData/XL1_R2.fq.gz |
| XL2 | XL2_R1 | RawData/XL2_R1.fq.gz |
Scan raw data
qc.pl: 使用fastqc和multiqc软件对raw data进行扫描,输入数据是 samples.fqpath.tsv,使用perl编程。
#!/usr/bin/perl
use warnings;
use strict;
use Getopt::Long;
my ($file, $real_dir, $out, $help, $version);
GetOptions(
"f|file:s" => \$file,
"d|real_dir:s" => \$real_dir,
"o|out:s" => \$out,
"h|help:s" => \$help,
"v|version" => \$version
);
&usage if(!defined $out);
# output
my $dir_qc = "$real_dir/result/00.quality/fastqc";
system "mkdir -p $dir_qc" unless(-d $dir_qc);
# script
my $dir_script = "$real_dir/result/script/00.quality/";
system "mkdir -p $dir_script" unless(-d $dir_script);
my @array_name;
open(IN, $file) or die "can't open $file\n";
open(OT, "> $out") or die "can't open $out\n";
<IN>;
while(<IN>){
chomp;
my @tmp = split("\t", $_);
if(-e $tmp[2]){
my $bash = join("", $dir_script, $tmp[1], ".fastqc.sh");
open(OT2, "> $bash") or die "can't open $bash\n";
print OT2 "fastqc -o $dir_qc --noextract $tmp[2]\n";
close(OT2);
print OT "sh $bash\n";
}
}
close(IN);
my $dir_mc = "$real_dir/result/00.quality/multiqc";
system "mkdir -p $dir_mc" unless(-d $dir_mc);
print OT "multiqc $dir_qc --outdir $dir_mc\n";
close(OT);
sub usage{
print <<USAGE;
usage:
perl $0 -f <file> -d <real_dir> -o <out>
options:
-f|file :[essential].
-d|real_dir :[essential].
-o|out :[essential].
USAGE
exit;
};
sub version {
print <<VERSION;
version: v1.1
update: 20201223 - 20201224
author: zouhua1\@outlook.com
VERSION
};
trim low quality reads and remove host DNA
kneaddata.pl:kneaddata内部自带过滤和比对软件
#!/usr/bin/perl
use warnings;
use strict;
use Getopt::Long;
my ($file, $real_dir, $out, $adapter, $help, $version);
GetOptions(
"f|file:s" => \$file,
"d|real_dir:s" => \$real_dir,
"a|adapt:s" => \$adapter,
"o|out:s" => \$out,
"h|help:s" => \$help,
"v|version" => \$version
);
&usage if(!defined $out);
my $dir = "$real_dir/result/01.kneaddata";
system "mkdir -p $dir" unless(-d $dir);
# script
my $dir_script = "$real_dir/result/script/01.kneaddata/";
system "mkdir -p $dir_script" unless(-d $dir_script);
open(IN, $file) or die "can't open $file";
my %file_name;
<IN>;
while(<IN>){
chomp;
my @tmp = split("\t", $_);
push (@{$file_name{$tmp[0]}}, $tmp[2]);
}
close(IN);
my ($fq1, $fq2);
open(OT, "> $out") or die "can't open $out\n";
foreach my $key (keys %file_name){
if (${$file_name{$key}}[0] =~ /R1/){
$fq1 = ${$file_name{$key}}[0];
}else{
$fq2 = ${$file_name{$key}}[0];
}
if (${$file_name{$key}}[1] =~ /R2/){
$fq2 = ${$file_name{$key}}[1];
}else{
$fq1 = ${$file_name{$key}}[1];
}
my $trim_opt = "ILLUMINACLIP:$adapter:2:40:15 SLIDINGWINDOW:4:20 MINLEN:50";
#if(-e $fq1 && -e $fq2){
my $bash = join("", $dir_script, $key, ".kneaddata.sh");
open(OT2, "> $bash") or die "can't open $bash\n";
print OT2 "kneaddata -i $fq1 -i $fq2 --output-prefix $key -o $dir -v -t 5 --remove-intermediate-output --trimmomatic /data/share/anaconda3/share/trimmomatic/ --trimmomatic-options \'$trim_opt\' --bowtie2-options \'--very-sensitive --dovetail\' -db /data/share/database/kneaddata_database/Homo_sapiens_Bowtie2_v0.1/Homo_sapiens\n";
close(OT2);
print OT "sh $bash\n";
#}
}
print OT "kneaddata_read_count_table --input $dir --output $dir/01kneaddata_sum.tsv\n";
close(OT);
sub usage{
print <<USAGE;
usage:
perl $0 -f <file> -d <real_dir> -o <out> -a <adapter>
options:
-f|file :[essential].
-d|real_dir :[essential].
-o|out :[essential].
-a|adapt :[essential].
USAGE
exit;
};
sub version {
print <<VERSION;
version: v1.1
update: 20201223 - 20201224
author: zouhua1\@outlook.com
VERSION
};
merge PE data
merge.pl:合并PE数据
#!/usr/bin/perl
use warnings;
use strict;
use Getopt::Long;
my ($file, $real_dir, $out, $help, $version);
GetOptions(
"f|file:s" => \$file,
"d|real_dir:s" => \$real_dir,
"o|out:s" => \$out,
"h|help:s" => \$help,
"v|version" => \$version
);
&usage if(!defined $out);
my $dir = "$real_dir/result/02.merge";
system "mkdir -p $dir" unless(-d $dir);
# script
my $dir_script = "$real_dir/result/script/02.merge/";
system "mkdir -p $dir_script" unless(-d $dir_script);
open(IN, $file) or die "can't open $file";
my %file_name;
<IN>;
while(<IN>){
chomp;
my @tmp = split("\t", $_);
push (@{$file_name{$tmp[0]}}, $tmp[2]);
}
close(IN);
my ($fq1, $fq2);
open(OT, "> $out") or die "can't open $out\n";
foreach my $key (keys %file_name){
$fq1 = join("", "./result/01.kneaddata/", $key, "_paired_1.fastq");
$fq2 = join("", "./result/01.kneaddata/", $key, "_paired_2.fastq");
#if(-e $fq1 && -e $fq2){
my $bash = join("", $dir_script, $key, ".merge.sh");
open(OT2, "> $bash") or die "can't open $bash\n";
print OT2 "fastp -i $fq1 -I $fq2 -h $dir/$key\_merge.html -j $dir/$key\_merge.json -m --merged_out $dir/$key\_merge.fastq.gz --failed_out $dir/$key\_failed.fastq.gz --include_unmerged --overlap_len_require 6 --overlap_diff_percent_limit 20 --detect_adapter_for_pe -5 -r -l 20 -y --thread 5\n";
close(OT2);
print OT "sh $bash\n";
#}
}
close(OT);
sub usage{
print <<USAGE;
usage:
perl $0 -f <file> -d <real_dir> -o <out>
options:
-f|file :[essential].
-d|real_dir :[essential].
-o|out :[essential].
USAGE
exit;
};
sub version {
print <<VERSION;
version: v1.1
update: 20201223 - 20201224
author: zouhua1\@outlook.com
VERSION
};
get profile matrix
humann.pl:获取功能等profile数据
#!/usr/bin/perl
use warnings;
use strict;
use Getopt::Long;
my ($file, $real_dir, $out, $help, $version);
GetOptions(
"f|file:s" => \$file,
"d|real_dir:s" => \$real_dir,
"o|out:s" => \$out,
"h|help:s" => \$help,
"v|version" => \$version
);
&usage if(!defined $out);
my $dir = "$real_dir/result/03.humann";
system "mkdir -p $dir" unless(-d $dir);
my $dir_log = "$real_dir/result/03.humann/log";
system "mkdir -p $dir_log" unless(-d $dir_log);
my $genefamilies = "$real_dir/result/03.humann/genefamilies";
system "mkdir -p $genefamilies" unless(-d $genefamilies);
my $pathabundance = "$real_dir/result/03.humann/pathabundance";
system "mkdir -p $pathabundance" unless(-d $pathabundance);
my $pathcoverage = "$real_dir/result/03.humann/pathcoverage";
system "mkdir -p $pathcoverage" unless(-d $pathcoverage);
my $dir_metaphlan = "$real_dir/result/03.humann/metaphlan";
system "mkdir -p $dir_metaphlan" unless(-d $dir_metaphlan);
# script
my $dir_script = "$real_dir/result/script/03.humann/";
system "mkdir -p $dir_script" unless(-d $dir_script);
open(IN, $file) or die "can't open $file";
my %file_name;
<IN>;
while(<IN>){
chomp;
my @tmp = split("\t", $_);
push (@{$file_name{$tmp[0]}}, $tmp[2]);
}
close(IN);
my ($fq);
open(OT, "> $out") or die "can't open $out\n";
foreach my $key (keys %file_name){
$fq = join("", "./result/02.merge/", $key, "_merge.fastq.gz");
#if($fq){
my $bash = join("", $dir_script, $key, ".humann.sh");
open(OT2, "> $bash") or die "can't open $bash\n";
print OT2 "humann --input $fq --output $dir --threads 10\n";
print OT2 "mv $dir/$key\_merge_humann_temp/$key\_merge_metaphlan_bugs_list.tsv $dir_metaphlan/$key\_metaphlan.tsv\n";
print OT2 "mv $dir/$key\_merge_humann_temp/$key\_merge.log $dir_log\n";
print OT2 "mv $dir/$key\_merge_genefamilies.tsv $genefamilies/$key\_genefamilies.tsv\n";
print OT2 "mv $dir/$key\_merge_pathabundance.tsv $pathabundance/$key\_pathabundance.tsv\n";
print OT2 "mv $dir/$key\_merge_pathcoverage.tsv $pathcoverage/$key\_pathcoverage.tsv\n";
print OT2 "rm -r $dir/$key\_merge_humann_temp/\n";
close(OT2);
print OT "sh $bash\n";
#}
}
close(OT);
sub usage{
print <<USAGE;
usage:
perl $0 -f <file> -d <real_dir> -o <out>
options:
-f|file :[essential].
-d|real_dir :[essential].
-o|out :[essential].
USAGE
exit;
};
sub version {
print <<VERSION;
version: v1.1
update: 20201223 - 20201225
author: zouhua1\@outlook.com
VERSION
};
metaphlan.pl:获取物种组成谱
#!/usr/bin/perl
use warnings;
use strict;
use Getopt::Long;
my ($file, $real_dir, $out, $help, $version);
GetOptions(
"f|file:s" => \$file,
"d|real_dir:s" => \$real_dir,
"o|out:s" => \$out,
"h|help:s" => \$help,
"v|version" => \$version
);
&usage if(!defined $out);
my $dir = "$real_dir/result/04.metaphlan";
system "mkdir -p $dir" unless(-d $dir);
# script
my $dir_script = "$real_dir/result/script/04.metaphlan/";
system "mkdir -p $dir_script" unless(-d $dir_script);
open(IN, $file) or die "can't open $file";
my %file_name;
<IN>;
while(<IN>){
chomp;
my @tmp = split("\t", $_);
push (@{$file_name{$tmp[0]}}, $tmp[2]);
}
close(IN);
my ($fq);
open(OT, "> $out") or die "can't open $out\n";
foreach my $key (keys %file_name){
$fq = join("", "./result/02.merge/", $key, "_merge.fastq.gz");
#if($fq){
my $bash = join("", $dir_script, $key, ".metaphlan.sh");
open(OT2, "> $bash") or die "can't open $bash\n";
print OT2 "metaphlan $fq --bowtie2out $dir/$key\_metagenome.bowtie2.bz2 --nproc 10 --input_type fastq -o $dir/$key\_metagenome.tsv --unknown_estimation -t rel_ab_w_read_stats\n";
close(OT2);
print OT "sh $bash\n";
#}
}
print OT "merge_metaphlan_tables.py $dir/*metagenome.tsv > $dir/merge_metaphlan.tsv\n";
print OT "Rscript /data/share/database/metaphlan_databases/calculate_unifrac.R $dir/merge_metaphlan.tsv /data/share/database/metaphlan_databases/mpa_v30_CHOCOPhlAn_201901_species_tree.nwk $dir/unifrac_merged_mpa3_profiles.tsv";
close(OT);
sub usage{
print <<USAGE;
usage:
perl $0 -f <file> -d <real_dir> -o <out>
options:
-f|file :[essential].
-d|real_dir :[essential].
-o|out :[essential].
USAGE
exit;
};
sub version {
print <<VERSION;
version: v1.0
update: 20201225 - 20201225
author: zouhua1\@outlook.com
VERSION
};
主程序
main.pl:生成所有的准备文件
#!/usr/bin/perl
use warnings;
use strict;
use Getopt::Long;
use FindBin qw($RealBin);
use Cwd 'abs_path';
my ($file, $adapter, $out, $help, $version);
GetOptions(
"f|file:s" => \$file,
"o|out:s" => \$out,
"a|adapter:s" => \$adapter,
"h|help:s" => \$help,
"v|version" => \$version
);
&usage if(!defined $out);
my $Bin = $RealBin;
my $cwd = abs_path;
# output
my $dir = "$cwd/result/";
system "mkdir -p $dir" unless(-d $dir);
########## output #########################################
# bash script per step
# combine all steps in one script
my $qc = join("", $dir, "Run.s1.qc.sh");
my $kneaddata = join("", $dir, "Run.s2.kneaddata.sh");
my $merge = join("", $dir, "Run.s3.merge.sh");
my $humann = join("", $dir, "Run.s4.humann.sh");
my $metaphlan = join("", $dir, "Run.s5.metaphlan.sh");
# scripts in bin
my $bin = "$Bin/bin";
my $s_qc = "$bin/qc.pl";
my $s_kneaddata = "$bin/kneaddata.pl";
my $s_merge = "$bin/merge.pl";
my $s_humann = "$bin/humann.pl";
my $s_metaphlan = "$bin/metaphlan.pl";
########## Steps in metagenomics pipeline #################
##################################################
# step1 reads quality scan
`perl $s_qc -f $file -d $cwd -o $qc`;
##################################################
# step2 filter and trim low quality reads;
# remove host sequence
`perl $s_kneaddata -f $file -d $cwd -a $adapter -o $kneaddata`;
##################################################
# step3 merge PE reads
`perl $s_merge -f $file -d $cwd -o $merge`;
##################################################
# step4 get function profile
`perl $s_humann -f $file -d $cwd -o $humann`;
##################################################
# step5 get taxonomy profile
`perl $s_metaphlan -f $file -d $cwd -o $metaphlan`;
open(OT, "> $out") or die "can't open $out\n";
print OT "sh $qc\nsh $kneaddata\nsh $merge\nsh $humann\nsh $metaphlan\n";
close(OT);
sub usage{
print <<USAGE;
usage:
perl $0 -f <file> -o <out> -a <adapter>
options:
-f|file :[essential].
-o|out :[essential].
-a|adapter :[essential].
USAGE
exit;
};
sub version {
print <<VERSION;
version: v1.1
update: 20201224 - 20201225
author: zouhua1\@outlook.com
VERSION
};
运行主程序:准备samples.fqpath.tsv和adapter.fa文件即可生成所有文件
perl main.pl -f samples.fqpath.tsv -a TruSeq2-PE.fa -o Run.all.sh
Run.all.sh 文件包含如下命令
sh /data/user/zouhua/pipeline/MetaGenomics_v2/result/Run.s1.qc.sh
sh /data/user/zouhua/pipeline/MetaGenomics_v2/result/Run.s2.kneaddata.sh
sh /data/user/zouhua/pipeline/MetaGenomics_v2/result/Run.s3.merge.sh
sh /data/user/zouhua/pipeline/MetaGenomics_v2/result/Run.s4.humann.sh
sh /data/user/zouhua/pipeline/MetaGenomics_v2/result/Run.s5.metaphlan.sh




![[论文阅读] 3D感知相关论文简单摘要](https://img-blog.csdnimg.cn/direct/9dd2899f189a43e6bc3df5cea7d42654.png)