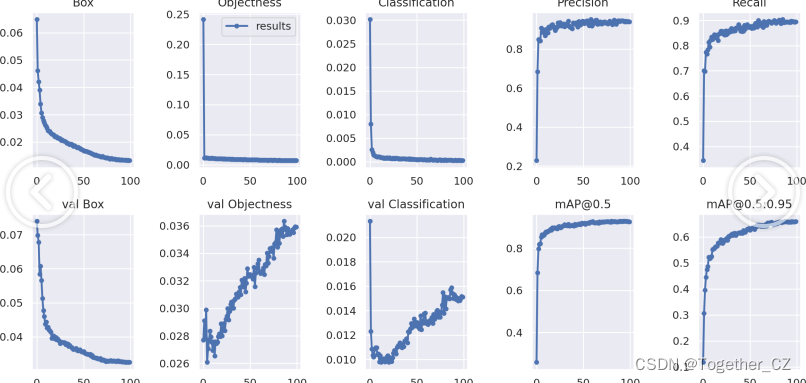
因为有同时反馈安装R包很慢或卡住。同事提供了一个安装R包的命令给我测试,在安装过程中复现报错信息,把下载慢或卡信的链接中的域名在防火墙中调整出口。
devtools::install_github("GreenleafLab/ArchR", ref="master", repos = BiocManager::repositories())下载安装miniconda
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
chmod +x Miniconda3-latest-Linux-x86_64.sh #给下载下来的安装文件添加执行权限
sudo bash Miniconda3-latest-Linux-x86_64.sh
一路Enter----yes—yes直到安装完成
进入conda环境
source ~/.bashrc
conda
国内换源
#全部清华园,备用
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/MindSpore
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/Paddle
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/auto
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/biobakery
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/bioconda
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/c4aarch64
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/caffe2
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/conda-forge
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/deepmodeling
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/dglteam
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/fastai
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/fermi
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/idaholab
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/intel
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/matsci
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/menpo/
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/mordred-descriptor
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/msys2
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/numba
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/ohmeta
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/omnia
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/peterjc123
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/plotly
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/psi4
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/pytorch
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/pytorch-lts
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/pytorch-test
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/pytorch3d
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/pyviz
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/qiime2
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/rapidsai
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/rdkit
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/simpleitk
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/stackless
conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/ursky
安装下R语言运行环境,安装R4.0.0(开始时安装R4.3.1提示不支持某个插件,具体不记得了)
conda search R

创建R4.0环境
conda create -n r4.0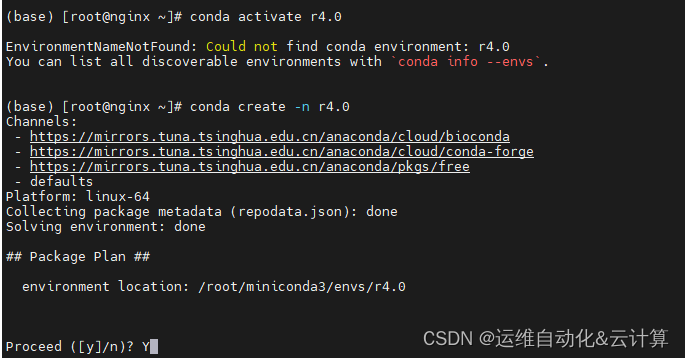
进入R4.0环境
conda activate r4.0安装R4.0
conda install R=4.0 -y 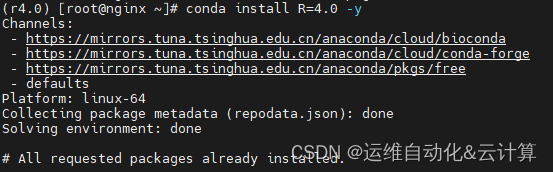
进入R程序
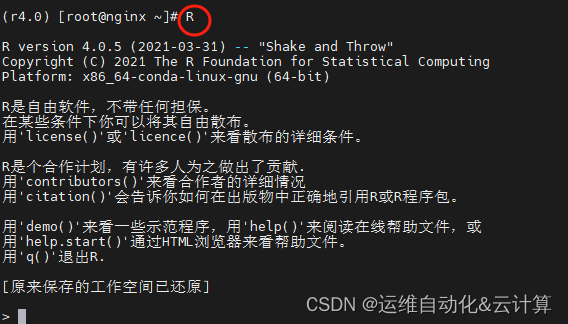
安装devtools
install.packages("devtools")安装R包报错:
1) grab failed: window not viewable.
Error in structure(.External(.C_dotTclObjv, objv), class = "tclObj") :
[tcl] grab failed: window not viewable.一般发生在远程linux系统中安装R包,是因为R调用窗口失败,解决方法是
chooseCRANmirror(graphics=FALSE)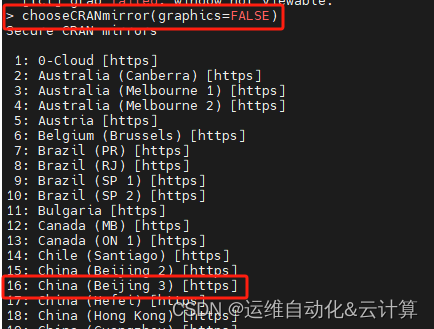
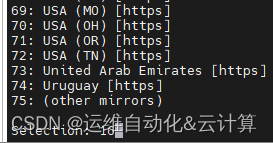
输入16回车,再运行一次安装命令。
install.packages("devtools")可能还会有下面出错信息
1: In install.packages("devtools", dependencies = TRUE) :
安装程序包‘systemfonts’时退出狀態的值不是0
2: In install.packages("devtools", dependencies = TRUE) :
安装程序包‘textshaping’时退出狀態的值不是0
3: In install.packages("devtools", dependencies = TRUE) :
安装程序包‘ragg’时退出狀態的值不是0
4: In install.packages("devtools", dependencies = TRUE) :
安装程序包‘pkgdown’时退出狀態的值不是0
5: In install.packages("devtools", dependencies = TRUE) :
安装程序包‘devtools’时退出狀態的值不是0解决办法:
1.install.packages('fontconfig')根据报错提示安装
yum install fontconfig-devel
2.install.packages('textshaping')根据报错提示安装
yum install -y harfbuzz-devel fribidi-devel
3.install.packages('ragg')根据报错提示安装
yum install -y freetype-devel libpng-devel libtiff-devel libjpeg-devel
4.处理完1.2.3再安装下面包已经不再提示错误了。
install.packages('pkgdown')
处理完再试试安装devtools.
在R中使用devtools安装R包
devtools::install_github("GreenleafLab/ArchR", ref="master", repos = BiocManager::repositories())例如生信分析常用到的Bioconductor,可使用BiocManager::install()来进行安装,例如安装DESeq2包
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("DESeq2")又报出错。
ERROR: dependencies ‘SummarizedExperiment’, ‘rhdf5’, ‘motifmatchr’, ‘chromVAR’, ‘uwot’, ‘Rsamtools’, ‘Biostrings’, ‘ComplexHeatmap’, ‘GenomicRanges’ are not available for package ‘ArchR’
* removing ‘/root/miniconda3/envs/r4.0/lib/R/library/ArchR’
使用pacman包来批量安装依赖包
install.packages("pacman")使用p_install函数,并通过packages参数传入要安装的包名列表:
pacman::p_install(packages = c("SummarizedExperiment", "rhdf5", "motifmatchr", "chromVAR", "uwot", "Rsamtools", "Biostrings", "ComplexHeatmap", "GenomicRanges"))出现ERROR: dependency ‘locfit’ is not available for package ‘DESeq2’ 的报错。此时往往是由于默认安装的locfit版本的问题导致的,所以通过指定低一点的版本(例如1.5-9.2)即可安装。
install.packages('https://cran.r-project.org/src/contrib/Archive/locfit/locfit_1.5-9.2.tar.gz',repos = NULL)出错揭示:
checking for gcc... no
checking for cc... no
checking for cl.exe... no
checking for clang... no
configure: error: in `/tmp/RtmpuOjLUv/R.INSTALLe06dd4f1f4256/XML':
configure: error: no acceptable C compiler found in $PATH
See `config.log' for more details
ERROR: configuration failed for package ‘XML’
* removing ‘/root/miniconda3/envs/r4.0/lib/R/library/XML’
* installing *source* package ‘BiocParallel’ ...
** using staged installation
checking for gcc... no
checking for cc... no
checking for cl.exe... no
configure: error: in `/tmp/RtmpsFlbCG/R.INSTALLe075b1e0ffb7b/BiocParallel':
configure: error: no acceptable C compiler found in $PATH
See `config.log' for more details
由于本机缺少gcc编译环境,通过yum安装gcc编译环境:
yum install -y gcc
参考文章:Miniconda3环境配置,换国内源_miniconda换源-CSDN博客
参考文章:没有人比我更懂R包安装 - 知乎
参考文章:DEseq2和edgeR的安装问题记录_deseq2安装报错-CSDN博客