Title
题目
Segment anything in medical images
在医学图像中进行任何分割
01
文献速递介绍
医学图像分割是临床实践中的关键组成部分,它促进了准确的诊断、治疗计划和疾病监测。
然而,现有的方法通常是针对特定的模态或疾病类型定制的,缺乏在医学图像分割任务的多样性谱系中的普遍适用性。在这里,我们介绍了MedSAM,这是一个旨在桥接这一差距的基础模型,通过使通用医学图像分割成为可能。该模型是在一个大规模的医学图像数据集上开发的,包含1,570,263个图像-掩码对,涵盖10种成像模态和30多种癌症类型。
我们在86个内部验证任务和60个外部验证任务上进行了全面评估,证明了该模型比模态专家模型具有更好的准确性和鲁棒性。通过在广泛的任务谱系中提供准确和高效的分割,MedSAM具有加速诊断工具发展和治疗计划个性化的重大潜力。
分割是医学成像分析中的一项基本任务,它涉及识别和勾画出各种医学图像中的感兴趣区域(ROI),例如器官、病变和组织。准确的分割对于许多临床应用至关重要,包括疾病诊断、治疗计划制定以及监测疾病进展。长期以来,手工分割一直是勾画解剖结构和病理区域的金标准,但这一过程耗时、劳动强度大,通常还需要高度的专业知识。半自动或全自动的分割方法可以显著减少所需的时间和劳动力,提高一致性,并使得大规模数据集的分析成为可能。
基于深度学习的模型在医学图像分割中显示出巨大的潜力,这归因于它们学习复杂图像特征和在多样化任务范围内提供准确分割结果的能力,从分割特定解剖结构到识别病理区域。然而,许多当前医学图像分割模型的一个显著限制是它们的任务特定性。这些模型通常被设计和训练用于特定的分割任务,当应用于新任务或不同类型的成像数据时,它们的性能可能会显著下降。这种缺乏通用性在临床实践中的广泛应用中构成了一个重大障碍。与此相反,自然图像分割领域的最近进展见证了分割基础模型的出现,如segment anything model(SAM)和Segment Everything Everywhere with Multi-modal prompts all at once,展示了在各种分割任务中的非凡多样性和性能。
METHOD
方法
Dataset curation and pre-processing
We curated a comprehensive dataset by collating images from publicly available medical image segmentation datasets, which were obtained from various sources across the internet, including the Cancer Imaging Archive (TCIA)34, Kaggle, Grand-Challenge, Scientific Data, CodaLab, and segmentation challenges in the Medical Image Computing and Computer Assisted Intervention Society (MICCAI). All the datasets provided segmentation annotations by human experts, which have been widely used in existing literature (Supplementary Table 1–4). We incorporated these annotations directly for both model development and validation. The original 3D datasets consisted of computed tomography (CT) and magnetic resonance (MR) images in DICOM, nrrd, or mhd formats.To ensure uniformity and compatibility with developing medical image deep learning models, we converted the images to the widely used NifTI format. Additionally, grayscale images (such as X-Ray and Ultrasound) as well as RGB images (including endoscopy, dermoscopy, fundus, and pathology images), were converted to the png format.
数据集策划和预处理
我们通过汇总公开可用的医学图像分割数据集中的图像,策划了一个全面的数据集,这些数据集从互联网上的各种来源获得,包括癌症影像档案(TCIA)、Kaggle、Grand-Challenge、Scientific Data、CodaLab以及医学图像计算和计算机辅助干预学会(MICCAI)的分割挑战。所有数据集都提供了人类专家的分割注释,这些注释在现有文献中已被广泛使用(补充表1-4)。我们直接将这些注释用于模型开发和验证。
原始的3D数据集由计算机断层扫描(CT)和磁共振(MR)图像组成,这些图像以DICOM、nrrd或mhd格式存在。为了确保与正在开发的医学图像深度学习模型的统一性和兼容性,我们将图像转换为广泛使用的NifTI格式。此外,灰度图像(如X射线和超声)以及RGB图像(包括内窥镜、皮肤镜、眼底和病理图像)被转换为png格式。
Results
结果
MedSAM: a foundation model for promptable medical image segmentation MedSAM aims to fulfill the role of a foundation model for universal medical image segmentation. A crucial aspect of constructing such a model is the capacity to accommodate a wide range of variations in imaging conditions, anatomical structures, and pathological condi tions. To address this challenge, we curated a diverse and large-scale medical image segmentation dataset with 1,570,263 medical image mask pairs, covering 10 imaging modalities, over 30 cancer types, and a multitude of imaging protocols (Fig. 1 and Supplementary Tables 1–4). This large-scale dataset allows MedSAM to learn a rich representation of medical images, capturing a broad spectrum of anatomies and lesions across different modalities. Figure 2a provides an overview of the distribution of images across different medical imaging modalities in the dataset, ranked by their total numbers. It is evident that computed tomography (CT), magnetic resonance ima ging (MRI), and endoscopy are the dominant modalities, reflecting their ubiquity in clinical practice. CT and MRI images provide detailed cross-sectional views of 3D body structures, making them indis pensable for non-invasive diagnostic imaging. Endoscopy, albeit more invasive, enables direct visual inspection of organ interiors, proving invaluable for diagnosing gastrointestinal and urological conditions. Despite the prevalence of these modalities, others such as ultrasound, pathology, fundus, dermoscopy, mammography, and optical coher ence tomography (OCT) also hold significant roles in clinical practice. The diversity of these modalities and their corresponding segmenta tion targets underscores the necessity for universal and effective segmentation models capable of handling the unique characteristics
associated with each modality.
MedSAM:一个用于可提示医学图像分割的基础模型
MedSAM 旨在充当通用医学图像分割的基础模型。构建这样一个模型的一个关键方面是能够适应成像条件、解剖结构和病理条件的广泛变化。为了应对这一挑战,我们策划了一个多样化且大规模的医学图像分割数据集,包含1,570,263个医学图像掩码对,涵盖10种成像模态、30多种癌症类型和众多成像协议(见图1及补充表1-4)。这个大规模数据集使MedSAM能够学习医学图像的丰富表示,捕捉不同模态下解剖结构和病变的广阔谱系。图2a提供了数据集中不同医学成像模态图像分布的概览,按照其总数进行排名。显然,计算机断层扫描(CT)、磁共振成像(MRI)和内窥镜是主导模态,反映了它们在临床实践中的普遍存在。CT和MRI图像提供了3D身体结构的详细横截面视图,使其成为非侵入性诊断成像不可或缺的部分。虽然内窥镜更具侵入性,但它能直接视察器官内部,对于诊断胃肠和泌尿系统疾病至关重要。
尽管这些模态很普遍,超声、病理、眼底、皮肤镜、乳腺X光和光学相干断层扫描(OCT)等其他模态在临床实践中也扮演着重要角色。这些模态的多样性及其相应的分割目标凸显了需要通用且有效的分割模型,这些模型能够处理与每种模态相关的独特特性的必要性。
Fig
图
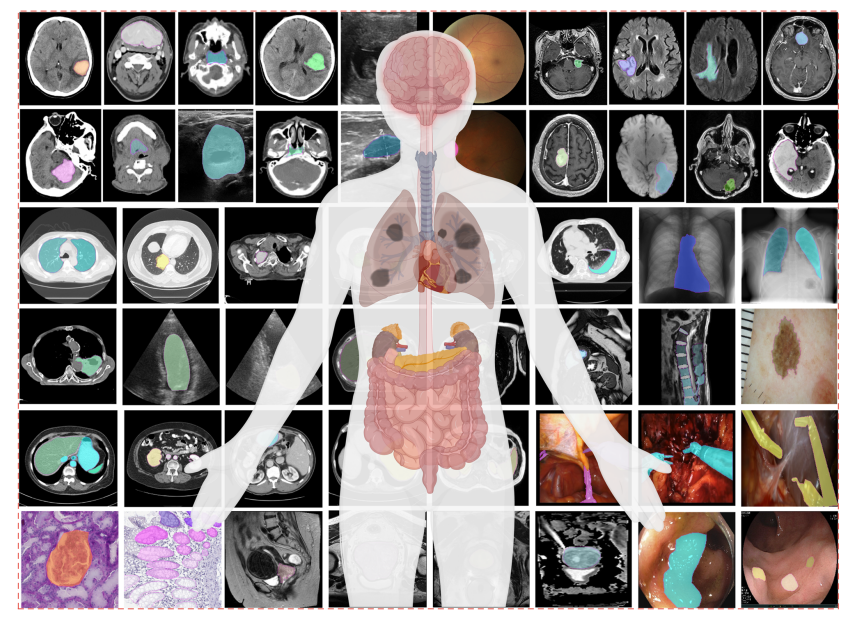
Fig. 1 | MedSAM is trained on a large-scale dataset that can handle diverse segmentation tasks. The dataset covers a variety of anatomical structures, pathological conditions, and medical imaging modalities. The magenta contours and mask overlays denote the expert annotations and MedSAM segmentation results, respectively.
图1 | MedSAM训练于一个大规模数据集,能够处理多样化的分割任务。该数据集涵盖了各种解剖结构、病理条件和医学成像模态。品红色轮廓和掩膜覆盖层分别代表专家标注和MedSAM分割结果。
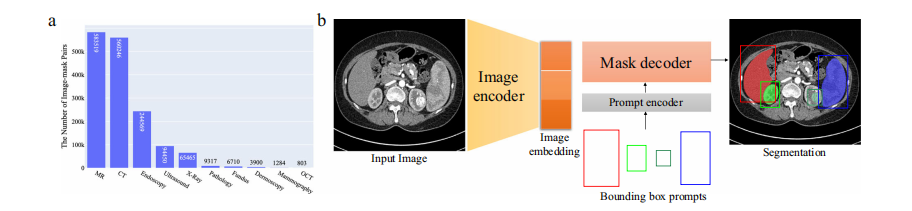
Fig. 2 | Overview of the modality distribution in the dataset and the network architecture. a The number of medical image-mask pairs in each modality. b MedSAM is a promptable segmentation method where users can use bounding boxes to specify the segmentation targets. Source data are provided as a Source Data file.
图2 | 数据集中模态分布的概览及网络架构。a 每种模态中医学图像-掩码对的数量。b MedSAM是一种可提示的分割方法,用户可以使用边界框来指定分割目标。源数据以源数据文件形式提供。
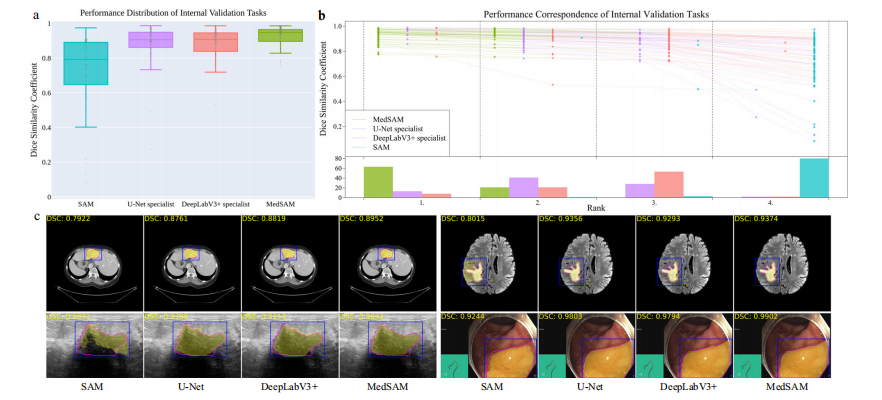
Fig. 3 | Quantitative and qualitative evaluation results on the internal validation set. a Performance distribution of 86 internal validation tasks in terms of median dice similarity coefficient (DSC) score. The center line within the box represents the median value, with the bottom and top bounds of the box deli neating the 25th and 75th percentiles, respectively. Whiskers are chosen to show the 1.5 of the interquartile range. Up-triangles denote the minima and down triangles denote the maxima. b Podium plots for visualizing the performance correspondence of 86 internal validation tasks. Upper part: each colored dot denotes the median DSC achieved with the respective method on one task. Dots corresponding to identical tasks are connected by a line. Lower part: bar charts represent the frequency of achieved ranks for each method. MedSAM ranks in the first place on most tasks. c Visualized segmentation examples on the internal validation set. The four examples are liver cancer, brain cancer, breast cancer, and polyp in computed tomography (CT), (Magnetic Resonance Imaging) MRI, ultra sound, and endoscopy images, respectively. Blue: bounding box prompts; Yellow: segmentation results. Magenta: expert annotations. Source data are provided as a Source Data file.
图3 | 内部验证集上的定量和定性评估结果。a 86个内部验证任务的性能分布,以中位数Dice相似系数(DSC)得分表示。盒图中的中心线代表中位值,盒子的底部和顶部边界分别界定了第25和第75百分位数。须表示为四分位数范围的1.5倍。向上的三角形表示最小值,向下的三角形表示最大值。b 颁奖台图,用于可视化86个内部验证任务的性能对应关系。上部:每个彩色点表示一项任务上使用相应方法所达到的中位DSC。相同任务对应的点通过线连接。下部:条形图代表每种方法达到的排名频率。MedSAM在大多数任务上排名第一。c 内部验证集上的可视化分割示例。四个示例分别是在计算机断层扫描(CT)、磁共振成像(MRI)、超声和内窥镜图像中的肝癌、脑癌、乳腺癌和息肉。蓝色:边界框提示;黄色:分割结果。品红色:专家标注。源数据以源数据文件形式提供。
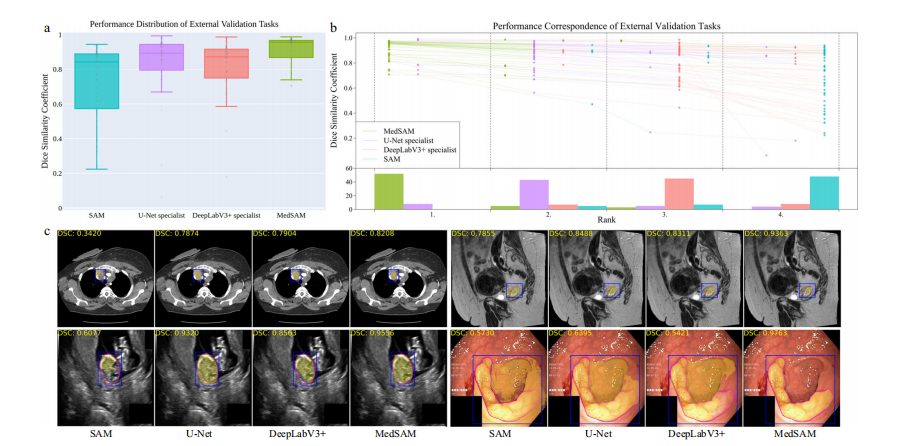
Fig. 4 | Quantitative and qualitative evaluation results on the external validation set. a Performance distribution of 60 external validation tasks in terms of median dice similarity coefficient (DSC) score. The center line within the box represents the median value, with the bottom and top bounds of the box deli neating the 25th and 75th percentiles, respectively. Whiskers are chosen to show the 1.5 of the interquartile range. Up-triangles denote the minima and down triangles denote the maxima. b Podium plots for visualizing the performance correspondence of 60 external validation tasks. Upper part: each colored dot denotes the median DSC achieved with the respective method on one task. Dots corresponding to identical tasks are connected by a line. Lower part: bar charts represent the frequency of achieved ranks for each method. MedSAM ranks in the first place on most tasks. c Visualized segmentation examples on the external validation set. The four examples are the lymph node, cervical cancer, fetal head, and polyp in CT, MR, ultrasound, and endoscopy images, respectively. Source data are provided as a Source Data file.
图4 | 外部验证集上的定量和定性评估结果。a 60个外部验证任务的性能分布,以中位数Dice相似系数(DSC)得分表示。盒图中的中心线代表中位值,盒子的底部和顶部边界分别界定了第25和第75百分位数。须表示为四分位数范围的1.5倍。向上的三角形表示最小值,向下的三角形表示最大值。b 颁奖台图,用于可视化60个外部验证任务的性能对应关系。上部:每个彩色点表示一项任务上使用相应方法所达到的中位DSC。相同任务对应的点通过线连接。下部:条形图代表每种方法达到的排名频率。MedSAM在大多数任务上排名第一。c 外部验证集上的可视化分割示例。四个示例分别是在CT、MR、超声和内窥镜图像中的淋巴结、宫颈癌、胎儿头部和息肉。源数据以源数据文件形式提供。
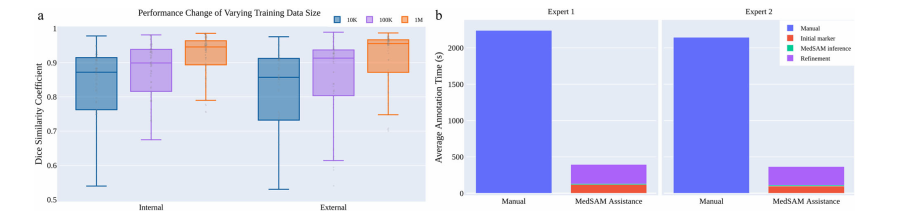
Fig. 5 | The effect of training dataset size and a user study of tumor annotation efficiency. a Scaling up the training image size to one million can significantly improve the model performance on both internal and external validation sets. b MedSAM can be used to substantially reduce the annotation time cost. Source data are provided as a Source Data file.
图5 | 训练数据集大小的影响及肿瘤标注效率的用户研究。a 将训练图像数量扩大到一百万可以显著提高模型在内部和外部验证集上的性能。b MedSAM可用于大幅减少标注时间成本。源数据以源数据文件形式提供。