> setwd("目录路径")
> library(vegan)
> library(picante)
> library(openxlsx)
> library(ggplot2)
> library(ggsci)
> otu <- read.xlsx("OTU.xlsx",rowNames = T)
> head(otu)
T1 T2 T3 T5 T6 T8 T9 N1 N2 N3 N4 N5
Abiotrophia 59231 34192 191 54 1355 268 0 0 0 4601 10372 382
Acholeplasma 0 0 0 2 0 0 0 0 0 0 0 0
Acidaminococcus 0 4 3 0 41 207 7 0 0 7 0 5
Acinetobacter 0 0 0 0 0 6 0 300 0 0 0 10
Actinobacillus 115 141 12 9 19 242 23 490 5 157 84 1706
Actinomyces 3069 2607 11200 18477 18230 14135 6988 13687 9790 6100 38734 17862
> otu <- t(otu) > head(otu) Abiotrophia Acholeplasma Acidaminococcus Acinetobacter Actinobacillus Actinomyces Actinotignum Aerococcus Aeromonas Aggregatibacter Aliarcobacter T1 59231 0 0 0 115 3069 0 13 0 830 0 T2 34192 0 4 0 141 2607 14 78 0 684 0 T3 191 0 3 0 12 11200 25 35 0 209 20 T5 54 2 0 0 9 18477 44 15 0 27 0 >>>>>>
> shannon=diversity(otu,"shannon") #计算香农指数
> simpson=diversity(otu,"simpson") #计算辛普森指数
> alpha=data.frame(shannon,simpson,check.names=T) #合并数据为数据框
> write.table(alpha,"alpha.csv",row.names=T,col.names=TRUE,sep=",") #保存数据
> df <- read.xlsx("a.xlsx") #在excel中整理了数据。
> head(df)
group shannon simpson
1 T 1.610562 0.7574733
2 T 1.808792 0.7744313
3 T 1.891420 0.7549007
4 T 1.244950 0.4794797
5 T 2.116994 0.7885307
6 T 2.266717 0.8429752
> df$group=factor(df$group)
> p<-ggplot(df, aes(x=group, y=shannon,fill=group)) +
geom_boxplot()+
scale_fill_brewer(palette="Set2")+
theme_classic()

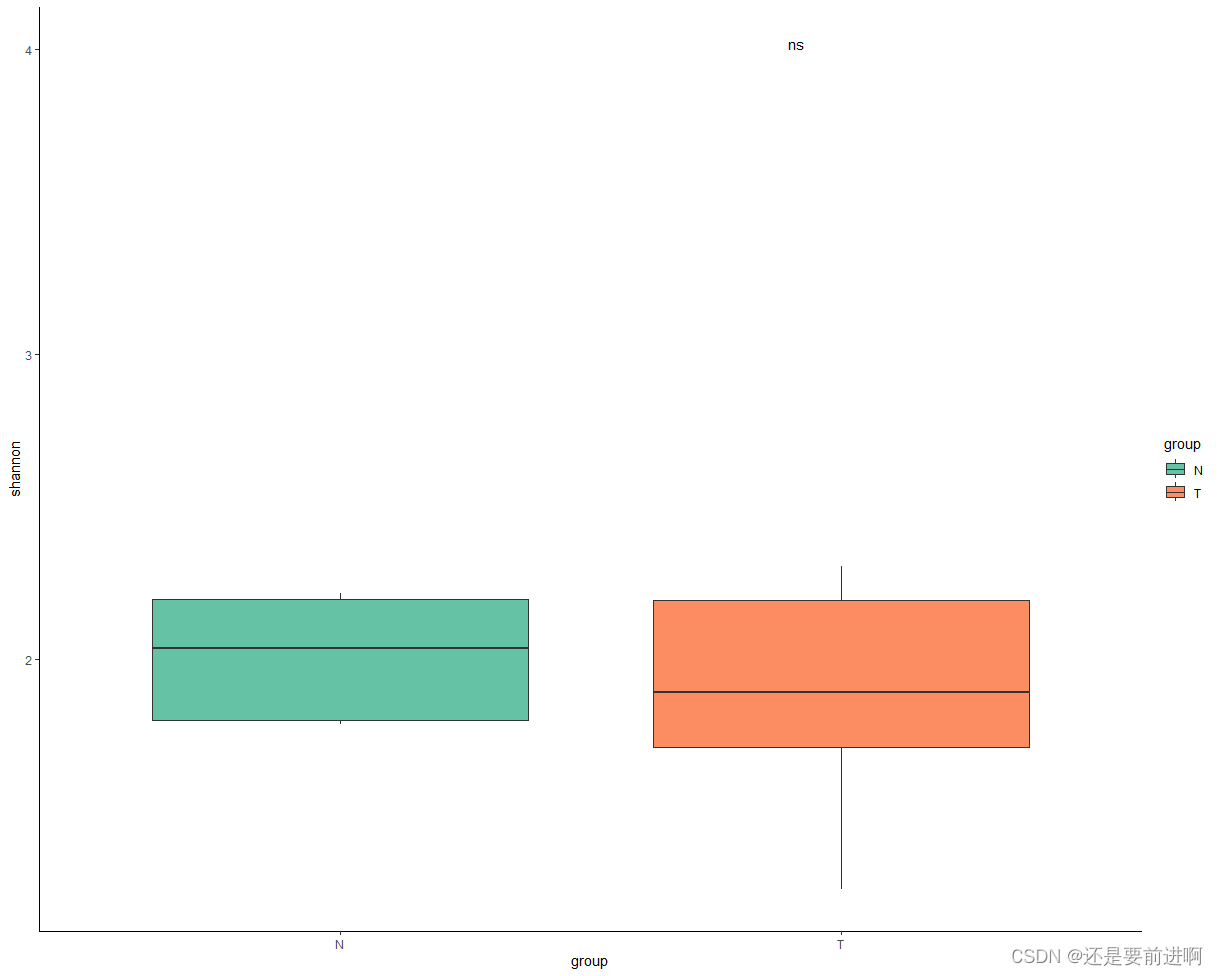
> p + stat_compare_means(method = "t.test",
label = "p.signif",
label.x = 1.9,
label.y = 4
) #加入两组差异比较,使用t.test。
> p+stat_compare_means(method = "anova",
label = "p.format",
label.x = 1.6,
label.y = 3.7
) # 加入两组差异比较,使用anova。
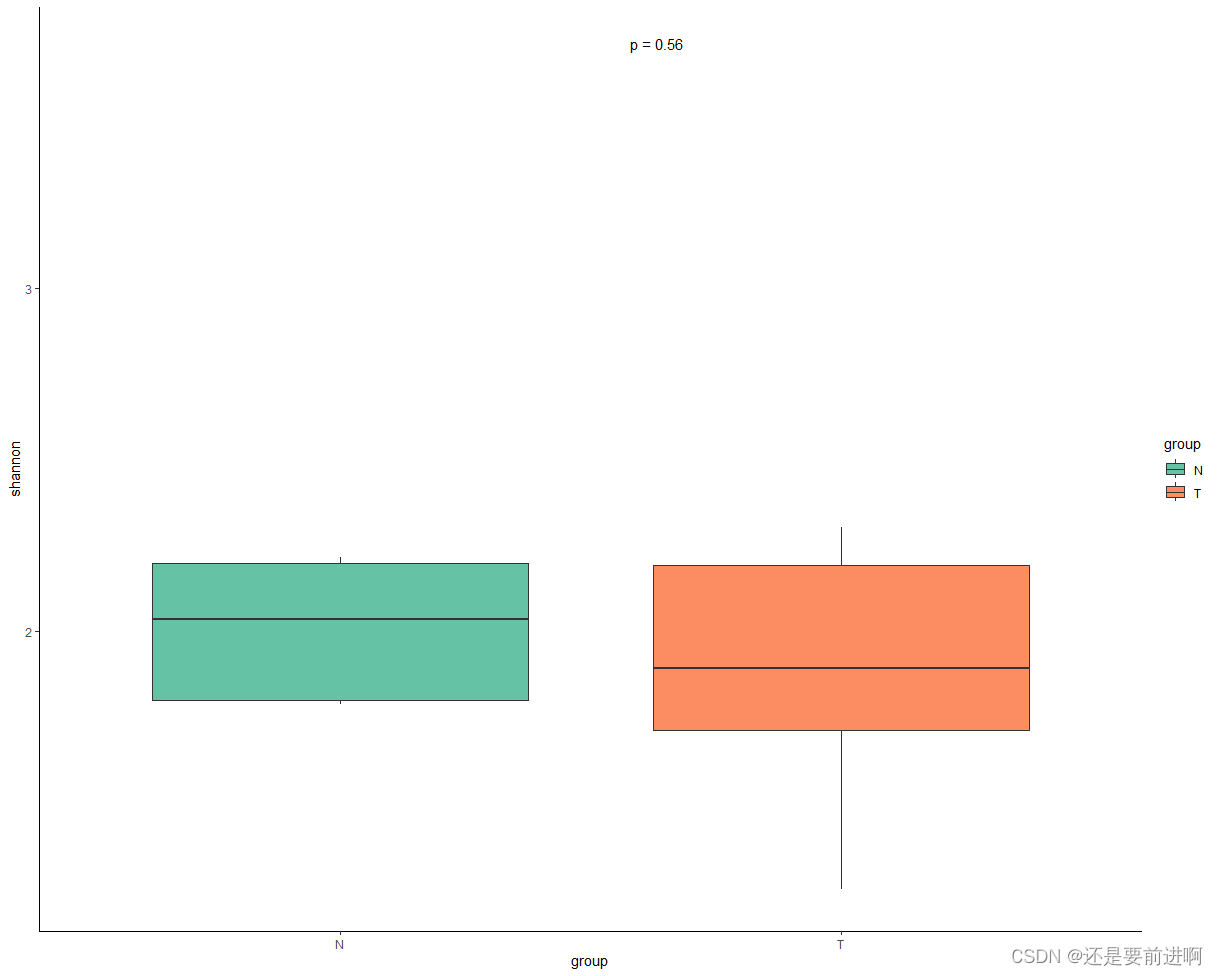
看来都没有显著差异。