内容如下:
1.外泌体和肝癌TCGA数据下载
2.数据格式整理
3.差异表达基因筛选
4.预后相关外泌体基因确定
5.拷贝数变异及突变图谱
6.外泌体基因功能注释
7.LASSO回归筛选外泌体预后模型
8.预后模型验证
9.预后模型鲁棒性分析
10.独立预后因素分析及与临床的相关性分析
11.列线图,ROC曲线,校准曲线,DCA曲线
12.外部数据集验证
13.外泌体模型与免疫的关系
14.外泌体模型与单细胞测序
############################## 02.数据格式整理 ###############################
下面进行数据格式整理,把TCGA肝癌数据进行基因去重复,并把肿瘤样本放在前面,正常样本放在后面,方便后续进行差异表达分析:
我们从网站上把FPKM格式的数据以及注释数据下载下来:
UCSC Xena![]() https://xenabrowser.net/datapages/?host=https%3A%2F%2Fgdc.xenahubs.net&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443
https://xenabrowser.net/datapages/?host=https%3A%2F%2Fgdc.xenahubs.net&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443
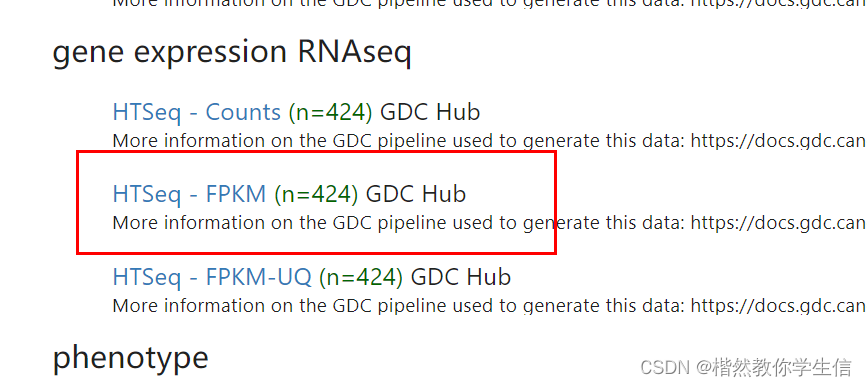
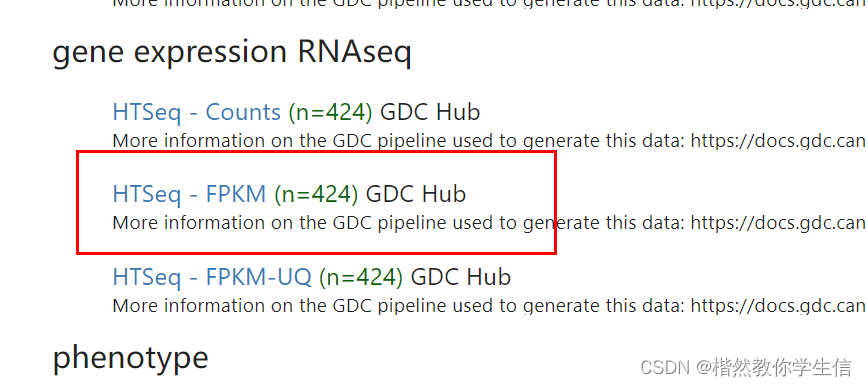

下面fpkm格式转换成TPM格式:
data <- read.csv("TCGA-LIHC.htseq_fpkm.tsv",header = T,sep = "\t")
data[1:5,1:5]
rownames(data) <- data$Ensembl_ID
data[1:5,1:5]
data <- data[,-1]
data <- 2^data-1
fpkmToTpm <- function(fpkm)
{
exp(log(fpkm) - log(sum(fpkm)) + log(1e6))
}
tpms <- apply(data,2,fpkmToTpm)
tpms[1:5,1:5]
colSums(tpms)
tpms <- log2(tpms+1)
write.csv(tpms,"LIHC_TPM.csv")
# > tpms[1:5,1:5]
# TCGA.DD.A4NG.01A TCGA.G3.AAV4.01A TCGA.2Y.A9H1.01A
#ENSG00000242268.2 0.000000 0.00000000 0.0000000
#ENSG00000270112.3 0.000000 0.03631085 0.0000000
#ENSG00000167578.15 1.549102 2.59344450 3.0610666
#ENSG00000273842.1 0.000000 0.00000000 0.0000000
#ENSG00000078237.5 2.058289 1.82597669 0.9323438
# TCGA.BC.A10Y.01A TCGA.K7.AAU7.01A
#ENSG00000242268.2 0.000000 0.48630273
#ENSG00000270112.3 0.000000 0.01992221
#ENSG00000167578.15 1.880890 3.07514645
#ENSG00000273842.1 0.000000 0.00000000
#ENSG00000078237.5 1.505011 2.55963800
这里还是geneid,我们需要换成genesymbol,因此要用到注释文件对基因进行注释:
dir()
data <- read.csv("LIHC_TPM.csv",header = T,sep = ",")
data[1:5,1:5]
annotation <- read.csv("gencode.v22.annotation.gene.probeMap",header = T,sep = "\t")
head(annotation)
match <- match(data$X,annotation$id)
head(match)
annotation <- annotation[match,]
head(annotation)
data[1:5,1:5]
identical(data$X,annotation$id)
data$X <- annotation$gene
data[1:5,1:5]
# > data[1:5,1:5]
# X TCGA.DD.A4NG.01A TCGA.G3.AAV4.01A TCGA.2Y.A9H1.01A TCGA.BC.A10Y.01A
#1 RP11-368I23.2 0.000000 0.00000000 0.0000000 0.000000
#2 RP11-742D12.2 0.000000 0.03631085 0.0000000 0.000000
#3 RAB4B 1.549102 2.59344450 3.0610666 1.880890
#4 AC104183.2 0.000000 0.00000000 0.0000000 0.000000
#5 C12orf5 2.058289 1.82597669 0.9323438 1.505011下面将肿瘤数据放在前面,正常数据放在后面,我们使用的正则表达式,这里的正则表达式只适用与TCGA的样本,下一节会讲一个万金油的办法:
注意TCGA样本的命名规则:
这里是0,表示肿瘤样本,是1表示正常样本。
rownames <- as.data.frame(data$X)
head(rownames)
names(rownames) <- "Symbol"
grep <- grep("^TCGA[.]([a-zA-Z0-9]{2})[.]([a-zA-Z0-9]{4})[.]([0][0-9][A-Z])",colnames(data))
length(grep)
grep
tumor <- data[,grep]
tumor[1:4,1:4]
grep1 <- grep("^TCGA[.]([a-zA-Z0-9]{2})[.]([a-zA-Z0-9]{4})[.]([1][0-9][A-Z])",colnames(data))
length(grep1)
grep1
normal <- data[,grep1]
normal[1:4,1:4]
data <- cbind(rownames,tumor,normal)
data[1:5,1:5]
library(limma)
rt=as.matrix(data)
rownames(rt) <- data[,1]
exp=rt[,2:ncol(rt)]
dimnames=list(rownames(exp),colnames(exp))
data33=matrix(as.numeric(as.matrix(exp)),nrow=nrow(exp),dimnames=dimnames)
## View(data33)
## BiocManager::install("limma")
library(limma)
dim(data33)
data33=avereps(data33)
dim(data33)
data33=data33[rowMeans(data33)>0,]
data33[1:5,1:5]
#> data33[1:5,1:5]
# TCGA.DD.A4NG.01A TCGA.G3.AAV4.01A TCGA.2Y.A9H1.01A TCGA.BC.A10Y.01A #TCGA.K7.AAU7.01A
#RP11-368I23.2 0.000000 0.00000000 0.0000000 0.000000 #0.48630273
#RP11-742D12.2 0.000000 0.03631085 0.0000000 0.000000 #0.01992221
#RAB4B 1.549102 2.59344450 3.0610666 1.880890 #3.07514645
#AC104183.2 0.000000 0.00000000 0.0000000 0.000000 #0.00000000
#C12orf5 2.058289 1.82597669 0.9323438 1.505011 #2.55963800
检查一下样本名字:
colnames(data33)
# > colnames(data33)
[1] "TCGA.DD.A4NG.01A" "TCGA.G3.AAV4.01A" "TCGA.2Y.A9H1.01A" "TCGA.BC.A10Y.01A"
[5] "TCGA.K7.AAU7.01A" "TCGA.BC.A10W.01A" "TCGA.DD.AACV.01A" "TCGA.DD.AAD3.01A"
[9] "TCGA.DD.A1EI.01A" "TCGA.DD.AAC9.01A" "TCGA.DD.AACT.01A" "TCGA.GJ.A6C0.01A"
[13] "TCGA.CC.5258.01A" "TCGA.DD.AADP.01A" "TCGA.DD.AACW.01A" "TCGA.ZS.A9CD.01A"
[17] "TCGA.UB.A7MF.01A" "TCGA.WX.AA46.01A" "TCGA.CC.A8HV.01A" "TCGA.DD.AADQ.01A"
[21] "TCGA.DD.AADB.01A" "TCGA.DD.A1EA.01A" "TCGA.5R.AA1C.01A" "TCGA.DD.A113.01A"
[25] "TCGA.2Y.A9H9.01A" "TCGA.CC.A7IE.01A" "TCGA.XR.A8TD.01A" "TCGA.DD.AAD2.01A"
[29] "TCGA.DD.AACY.01A" "TCGA.CC.A7IF.01A" "TCGA.DD.AACD.01A" "TCGA.ED.A4XI.01A""
[185] "TCGA.WQ.AB4B.01A" "TCGA.CC.A123.01A" "TCGA.YA.A8S7.01A" "TCGA.2Y.A9H2.01A"
[189] "TCGA.5C.A9VG.01A" "TCGA.CC.A3M9.01A" "TCGA.EP.A2KA.01A" "TCGA.DD.AADW.01A"
[193] "TCGA.DD.AAED.01A" "TCGA.LG.A9QD.01A" "TCGA.DD.AADV.01A" "TCGA.G3.A3CH.01A"
[197] "TCGA.G3.AAV1.01A" "TCGA.ZS.A9CG.01A" "TCGA.DD.AAW3.01A" "TCGA.ED.A7PY.01A"
[201] "TCGA.ZP.A9D2.01A" "TCGA.G3.A25Y.01A" "TCGA.G3.A3CI.01A" "TCGA.K7.A5RG.01A"
[381] "TCGA.DD.A39V.11A" "TCGA.BC.A110.11A" "TCGA.DD.A3A1.11A" "TCGA.DD.A1EB.11A"
[385] "TCGA.DD.A1EI.11A" "TCGA.BC.A10R.11A" "TCGA.DD.A1EH.11A" "TCGA.DD.A11C.11A"
[389] "TCGA.DD.A1EJ.11A" "TCGA.DD.A3A4.11A" "TCGA.BC.A10Y.11A" "TCGA.DD.A113.11A"
[393] "TCGA.DD.A1EG.11A" "TCGA.G3.A3CH.11A" "TCGA.BC.A10X.11A" "TCGA.DD.A39Z.11A"
[397] "TCGA.BC.A10W.11A" "TCGA.BD.A3EP.11A" "TCGA.FV.A3I0.11A" "TCGA.DD.A3A8.11A"
[401] "TCGA.DD.A116.11A" "TCGA.FV.A23B.11A" "TCGA.FV.A3I1.11A" "TCGA.BC.A10Q.11A"
[405] "TCGA.DD.A11D.11A" "TCGA.FV.A2QR.11A" "TCGA.DD.A11A.11A" "TCGA.DD.A3A2.11A"
[409] "TCGA.BD.A2L6.11A" "TCGA.EP.A12J.11A" "TCGA.DD.A3A6.11A" "TCGA.EP.A26S.11A"
[413] "TCGA.DD.A118.11A" "TCGA.DD.A119.11A" "TCGA.DD.A1EE.11A" "TCGA.BC.A216.11A"
[417] "TCGA.BC.A10Z.11A" "TCGA.FV.A3R2.11A" "TCGA.ES.A2HT.11A" "TCGA.DD.A114.11A"
[421] "TCGA.BC.A10T.11A" "TCGA.EP.A3RK.11A" "TCGA.BC.A10U.11A" "TCGA.DD.A11B.11A"可以看到,肿瘤样本在前面,正常样本在后面,读出数据以便下次使用:
write.csv(data33,"TCGA-LIHC.csv")
下一节进行差异表达分析。