引言
最初只是想把Doccano标注的数据集转换成BIO(类似conll2003数据集)的标注格式;
按照PR的修改意见实现了修改,但是本人不建议这么做;
应该随着Doccano的升级,Doccano的导出格式发生了变化,而原来的doccano-transformer还停留在2022年
摘要
可先阅读一下教程:【已解决】关于如何将Doccano标注的文本转换成NER模型可以直接处理的CoNLL 2003格式
装包:pip install doccano-transformer
报错信息
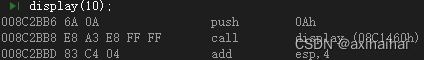
运行下述程序后,会报错
from doccano_transformer.datasets import NERDataset
from doccano_transformer.utils import read_jsonl
dataset = read_jsonl(filepath='NER.jsonl', dataset=NERDataset, encoding='utf-8')
gen=dataset.to_conll2003(tokenizer=str.split)
file_name="CoNLL.txt"
with open(file_name, "w", encoding = "utf-8") as file:
for item in gen:
file.write(item["data"] + "\n")
报错信息如下:
l In[18], line 1
----> 1 from doccano_transformer.datasets import NERDataset
2 from doccano_transformer.utils import read_jsonl
File ~/anaconda3/envs/nlp/lib/python3.9/site-packages/doccano_transformer/datasets.py:5
2 import json
3 from typing import Any, Callable, Iterable, Iterator, List, Optional, TextIO
----> 5 from doccano_transformer.examples import Example, NERExample
8 class Dataset:
9 def __init__(
10 self,
11 filepath: str,
12 encoding: Optional[str] = 'utf-8',
13 transformation_func: Optional[Callable[[TextIO], Iterable[Any]]] = None
14 ) -> None:
File ~/anaconda3/envs/nlp/lib/python3.9/site-packages/doccano_transformer/examples.py:4
1 from collections import defaultdict
2 from typing import Callable, Iterator, List, Optional
----> 4 from spacy.gold import biluo_tags_from_offsets
6 from doccano_transformer import utils
9 class Example:
ModuleNotFoundError: No module named 'spacy.gold'
修复bug
根据该GitHub doccano_transformer项目的github issues和pr 给出的信息修复该bug:
要修改doccano_transformer/examples.py源码文件;
根据报错信息,确定example.py文件所在目录
File ~/anaconda3/envs/nlp/lib/python3.9/site-packages/doccano_transformer/datasets.py:5
根据报错信息,知道笔者的examples.py路径如下:
(每人的所在文件夹不同,请自行修改)
~/anaconda3/envs/nlp/lib/python3.9/site-packages/doccano_transformer/examples.py
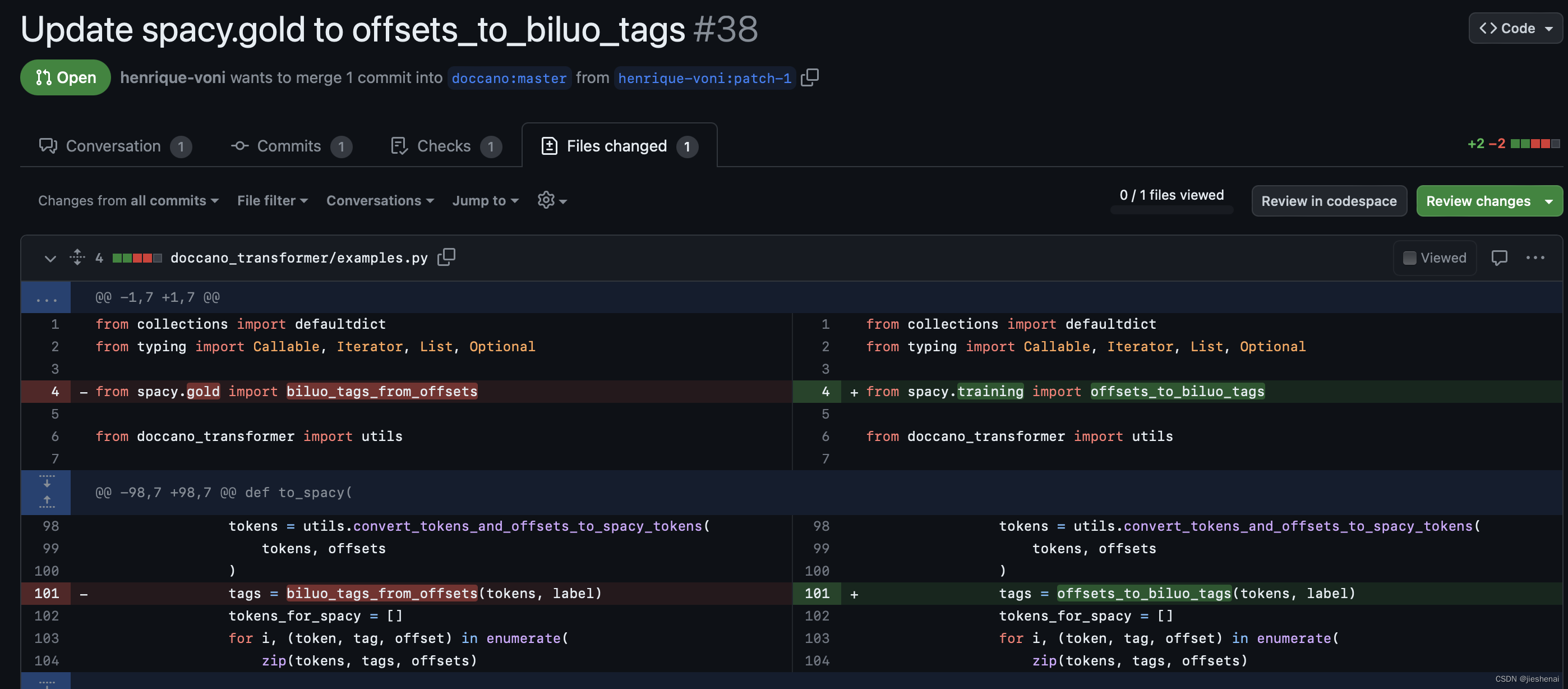
按照图片所示内容进行修改即可:
-
修改点 1
原始代码:
from spacy.gold import biluo_tags_from_offsets修改成:
from spacy.training import offsets_to_biluo_tags -
修改点 2
原始代码:
tags = biluo_tags_from_offsets(tokens, label)修改成:
tags = offsets_to_biluo_tags(tokens, label)
修改完上述代码,重新运行代码就不会报错了;
相关阅读
- BIO序列提取实体(NER命名实体识别)
该文把BIO标注的数据,转成下述格式:{'string': '我是李明,我爱中国,我来自呼和浩特', 'entities': [{'word': '中国', 'type': 'loc'}, {'word': '呼和浩特', 'type': 'loc'}]}
参考资料
- [1] github issues https://github.com/doccano/doccano-transformer/issues/35
- [2] 该bug的PR参考 https://github.com/doccano/doccano-transformer/pull/38/files