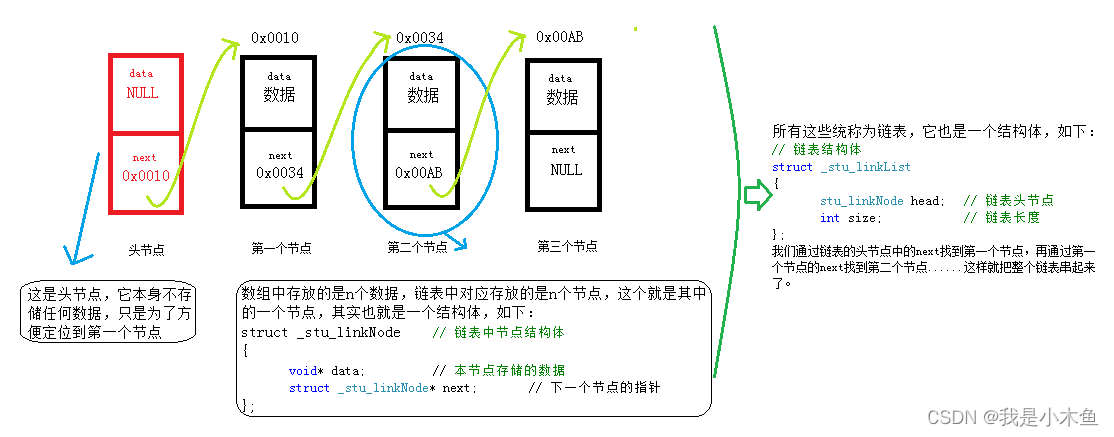
BikeDNA(六)参考数据的内在分析2
1.数据完整性
见链接
2.网络拓扑结构
见链接
3.网络组件
断开连接的组件不共享任何元素(节点/边)。 换句话说,不存在可以从一个断开连接的组件通向另一组件的网络路径。 如上所述,大多数现实世界的自行车基础设施网络确实由许多断开连接的组件组成(Natera Orozco et al., 2020) 。 然而,当两个断开的组件彼此非常接近时,这可能是边缘缺失或另一个数字化错误的迹象。
方法
首先,在“return_components”的帮助下,获得网络所有(断开连接的)组件的列表。 打印组件总数,并以不同颜色绘制所有组件以进行视觉分析。 接下来,绘制组件大小分布(组件按其包含的网络长度排序),然后绘制最大连接组件的图。
解释
与之前的许多分析步骤一样,该领域的知识对于正确解释成分分析至关重要。 鉴于数据准确地代表了实际的基础设施,较大的组件表示连贯的网络部分,而较小的组件表示分散的基础设施(例如,沿着街道的一条自行车道,不连接到任何其他自行车基础设施)。 大量彼此邻近的断开组件表明数字化错误或丢失数据。
3.1 断开的组件
ref_components = eval_func.return_components(ref_graph_simplified)
print(
f"The network in the study area has {len(ref_components)} disconnected components."
)
The network in the study area has 204 disconnected components.
# Plot disconnected components
set_renderer(renderer_map)
# set seed for colors
np.random.seed(42)
# generate enough random colors to plot all components
randcols = np.random.rand(len(ref_components), 3)
randcols[0, :] = col_to_rgb(pdict['ref_base'])
fig, ax = plt.subplots(1, 1, figsize=pdict["fsmap"])
ax.set_title(f"{area_name}: {reference_name} disconnected components")
ax.set_axis_off()
for j, c in enumerate(ref_components):
if len(c.edges) > 0:
edges = ox.graph_to_gdfs(c, nodes=False)
edges.plot(ax=ax, color=randcols[j])
cx.add_basemap(ax=ax, crs=study_crs, source=cx_tile_2)
cx.add_attribution(ax=ax, text=f"(C) {reference_name}")
txt = ax.texts[-1]
txt.set_position([1,0.00])
txt.set_ha('right')
txt.set_va('bottom')
plot_func.save_fig(fig, ref_results_static_maps_fp + "all_components_reference")

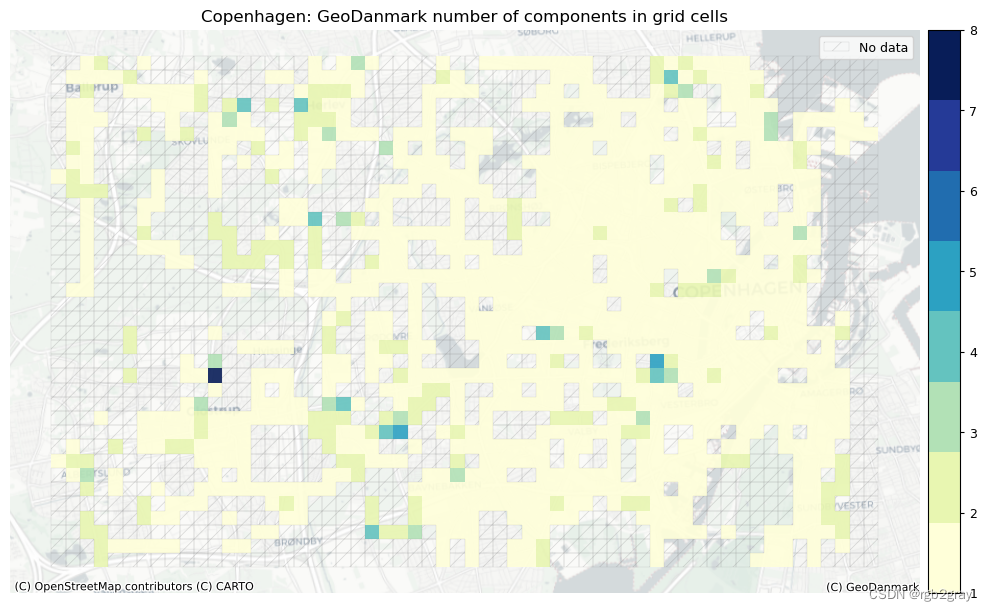
3.2 每个网格单元的组件
# Assign component ids to grid
grid = eval_func.assign_component_id_to_grid(
ref_edges_simplified,
ref_edges_simp_joined,
ref_components,
grid,
prefix="ref",
edge_id_col="edge_id",
)
fill_na_dict = {"component_ids_ref": ""}
grid.fillna(value=fill_na_dict, inplace=True)
grid["component_count_ref"] = grid.component_ids_ref.apply(lambda x: len(x))
# Plot number of components per grid cell
set_renderer(renderer_map)
fig, ax = plt.subplots(1, 1, figsize=pdict["fsmap"])
ncolors = grid["component_count_ref"].max()
from mpl_toolkits.axes_grid1 import make_axes_locatable
divider = make_axes_locatable(ax)
cax = divider.append_axes("right", size="3.5%", pad="1%")
mycm = cm.get_cmap(pdict["seq"], ncolors)
grid[grid.component_count_ref>0].plot(
cax=cax,
ax=ax,
column="component_count_ref",
legend=True,
legend_kwds={'ticks': list(range(1, ncolors+1))},
cmap=mycm,
alpha=pdict["alpha_grid"],
)
# add no data patches
grid[grid["count_ref_edges"].isnull()].plot(
cax=cax,
ax=ax,
facecolor=pdict["nodata_face"],
edgecolor=pdict["nodata_edge"],
linewidth= pdict["line_nodata"],
hatch=pdict["nodata_hatch"],
alpha=pdict["alpha_nodata"],
)
ax.legend(handles=[nodata_patch], loc="upper right")
cx.add_basemap(ax=ax, crs=study_crs, source=cx_tile_2)
cx.add_attribution(ax=ax, text=f"(C) {reference_name}")
txt = ax.texts[-1]
txt.set_position([1,0.00])
txt.set_ha('right')
txt.set_va('bottom')
ax.set_title(area_name + f": {reference_name} number of components in grid cells")
ax.set_axis_off()
plot_func.save_fig(fig, ref_results_static_maps_fp + f"number_of_components_in_grid_cells_reference")
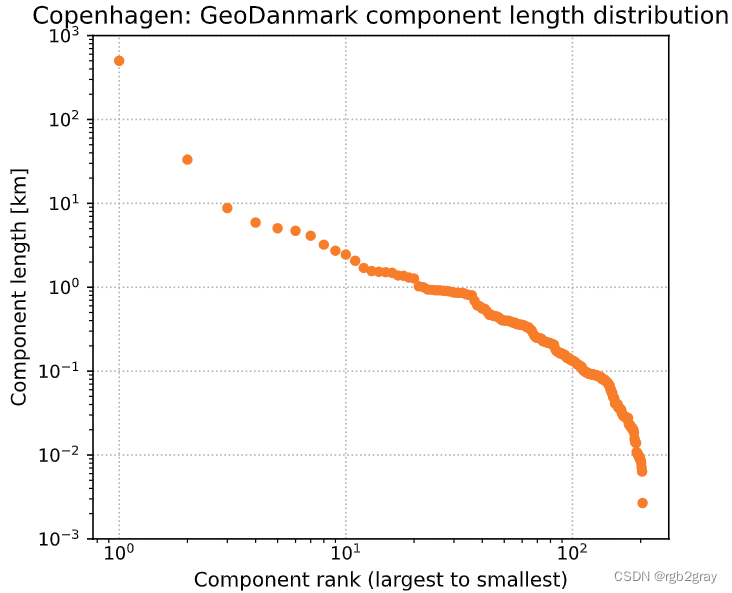
3.3 组件长度分布
所有网络组件长度的分布可以在所谓的 Zipf 图 中可视化,该图按等级对每个组件的长度进行排序,在左侧显示最大组件的长度,然后是第二大组件的长度,依此类推,直到 右侧最小组件的长度。 当 Zipf 图遵循 双对数比例 中的直线时,这意味着找到小的不连续组件的机会比传统分布的预期要高得多 (Clauset et al., 2009)。 这可能意味着网络没有合并,只有分段或随机添加 (Szell et al., 2022),或者数据本身存在许多间隙和拓扑错误,导致小的断开组件。
但是,也可能发生最大的连通分量(图中最左边的标记,等级为 1 0 0 10^0 100)是明显的异常值,而图的其余部分则遵循不同的形状。 这可能意味着在基础设施层面,大部分基础设施已连接到一个大型组件,并且数据反映了这一点 - 即数据在很大程度上没有受到间隙和缺失链接的影响。
自行车网络也可能介于两者之间,有几个大型组件作为异常值。
# Zipf plot of component lengths
set_renderer(renderer_plot)
components_length = {}
for i, c in enumerate(ref_components):
c_length = 0
for (u, v, l) in c.edges(data="length"):
c_length += l
components_length[i] = c_length
components_df = pd.DataFrame.from_dict(components_length, orient="index")
components_df.rename(columns={0: "component_length"}, inplace=True)
fig = plt.figure(figsize=pdict["fsbar_small"])
axes = fig.add_axes([0, 0, 1, 1])
axes.set_axisbelow(True)
axes.grid(True,which="major",ls="dotted")
yvals = sorted(list(components_df["component_length"] / 1000), reverse = True)
axes.scatter(
x=[i+1 for i in range(len(components_df))],
y=yvals,
s=18,
color=pdict["ref_base"],
)
axes.set_ylim(ymin=10**math.floor(math.log10(min(yvals))), ymax=10**math.ceil(math.log10(max(yvals))))
axes.set_xscale("log")
axes.set_yscale("log")
axes.set_ylabel("Component length [km]")
axes.set_xlabel("Component rank (largest to smallest)")
axes.set_title(area_name+": " + f"{reference_name} component length distribution")
plot_func.save_fig(fig, ref_results_plots_fp + "component_length_distribution_reference")
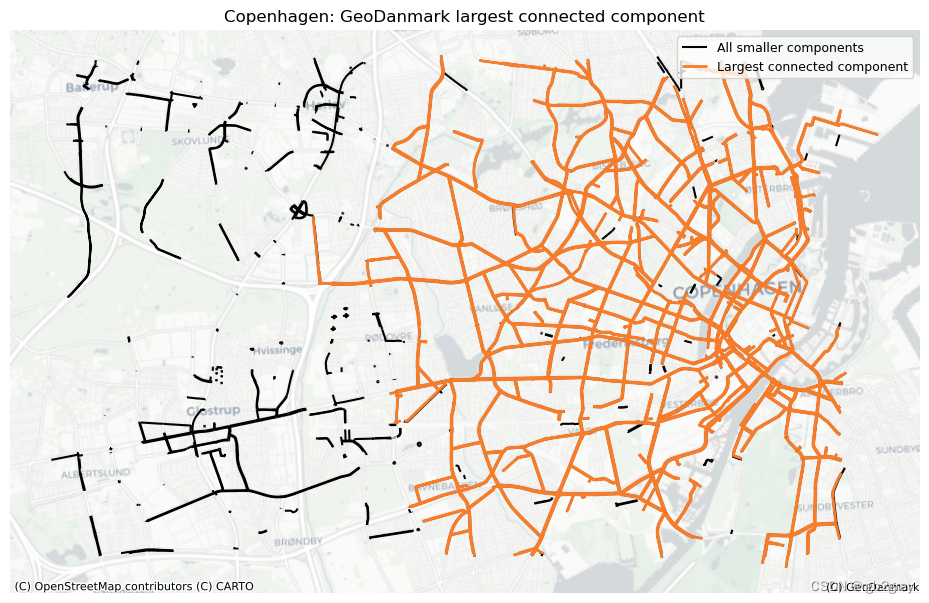
largest_cc = max(ref_components, key=len)
largest_cc_length = 0
for (u, v, l) in largest_cc.edges(data="length"):
largest_cc_length += l
largest_cc_pct = largest_cc_length / components_df["component_length"].sum() * 100
print(
f"The largest connected component contains {largest_cc_pct:.2f}% of the network length."
)
lcc_edges = ox.graph_to_gdfs(
G=largest_cc, nodes=False, edges=True, node_geometry=False, fill_edge_geometry=False
)
# Export to GPKG
lcc_edges[["edge_id", "geometry"]].to_file(
ref_results_data_fp + "largest_connected_component.gpkg"
)
The largest connected component contains 80.04% of the network length.
3.4 最大连通分量
# Plot of largest connected component
set_renderer(renderer_map)
fig, ax = plt.subplots(1, 1, figsize=pdict["fsmap"])
ref_edges_simplified.plot(ax=ax, color = pdict["base"], linewidth = 1.5, label = "All smaller components")
lcc_edges.plot(ax=ax, color=pdict["ref_base"], linewidth = 2, label = "Largest connected component")
grid.plot(ax=ax,alpha=0)
ax.set_axis_off()
ax.set_title(area_name + f": {reference_name} largest connected component")
ax.legend()
cx.add_basemap(ax=ax, crs=study_crs, source=cx_tile_2)
cx.add_attribution(ax=ax, text=f"(C) {reference_name}")
txt = ax.texts[-1]
txt.set_position([1,0.00])
txt.set_ha('right')
txt.set_va('bottom')
plot_func.save_fig(fig, ref_results_static_maps_fp + f"largest_conn_comp_reference")
# Save plot without basemap for potential report titlepage
set_renderer(renderer_map)
fig, ax = plt.subplots(1, 1, figsize=pdict["fsmap"])
ref_edges_simplified.plot(ax=ax, color = pdict["base"], linewidth = 1.5, label = "Disconnected components")
lcc_edges.plot(ax=ax, color=pdict["ref_base"], linewidth = 2, label = "Largest connected component")
ax.set_axis_off()
plot_func.save_fig(fig, ref_results_static_maps_fp + "titleimage",plot_res="high")
plt.close()
3.5 缺少链接
在组件之间潜在缺失链接的图中,将绘制与另一个组件上的边的指定距离内的所有边。 断开的边缘之间的间隙用标记突出显示。
因此,该地图突出显示了边缘,尽管这些边缘彼此非常接近,但它们是断开连接的,因此不可能在边缘之间的自行车基础设施上骑自行车。
在分析组件之间潜在的缺失链接时,用户必须定义两个组件之间的距离被认为足够低以至于怀疑数字化错误的阈值。
# DEFINE MAX BUFFER DISTANCE BETWEEN COMPONENTS CONSIDERED A GAP/MISSING LINK
component_min_distance = 10
assert isinstance(component_min_distance, int) or isinstance(
component_min_distance, float
), print("Setting must be integer or float value!")
print(f"Running analysis with component distance threshold of {component_min_distance} meters.")
Running analysis with component distance threshold of 10 meters.
component_gaps = eval_func.find_adjacent_components(
components=ref_components,
buffer_dist=component_min_distance,
crs=study_crs,
edge_id="edge_id",
)
component_gaps_gdf = gpd.GeoDataFrame.from_dict(
component_gaps, orient="index", geometry="geometry", crs=study_crs
)
edge_ids = set(
component_gaps_gdf["edge_id" + "_left"].to_list()
+ component_gaps_gdf["edge_id" + "_right"].to_list()
)
edge_ids = [int(i) for i in edge_ids]
edges_with_gaps = ref_edges_simplified.loc[ref_edges_simplified.edge_id.isin(edge_ids)]
# Save to csv
pd.DataFrame(edge_ids, columns=["edge_id"]).to_csv(
ref_results_data_fp + f"component_gaps_edges_{component_min_distance}.csv",
index=False,
)
component_gaps_gdf.to_file(
ref_results_data_fp + f"component_gaps_centroids_{component_min_distance}.gpkg"
)
# Interactive plot of adjacent components
if len(component_gaps) > 0:
simplified_edges_folium = plot_func.make_edgefeaturegroup(
gdf=ref_edges_simplified,
mycolor=pdict["ref_base"],
myweight=pdict["line_base"],
nametag="All edges",
show_edges=True,
)
component_issues_edges_folium = plot_func.make_edgefeaturegroup(
gdf=edges_with_gaps,
mycolor=pdict["ref_emp"],
myweight=pdict["line_emp"],
nametag="Adjacent disconnected edges",
show_edges=True,
)
component_issues_gaps_folium = plot_func.make_markerfeaturegroup(
gdf=component_gaps_gdf, nametag="Component gaps", show_markers=True
)
m = plot_func.make_foliumplot(
feature_groups=[
simplified_edges_folium,
component_issues_edges_folium,
component_issues_gaps_folium,
],
layers_dict=folium_layers,
center_gdf=ref_nodes_simplified,
center_crs=ref_nodes_simplified.crs,
)
bounds = plot_func.compute_folium_bounds(ref_nodes_simplified)
m.fit_bounds(bounds)
m.save(ref_results_inter_maps_fp + f"component_gaps_{component_min_distance}_reference.html")
display(m)
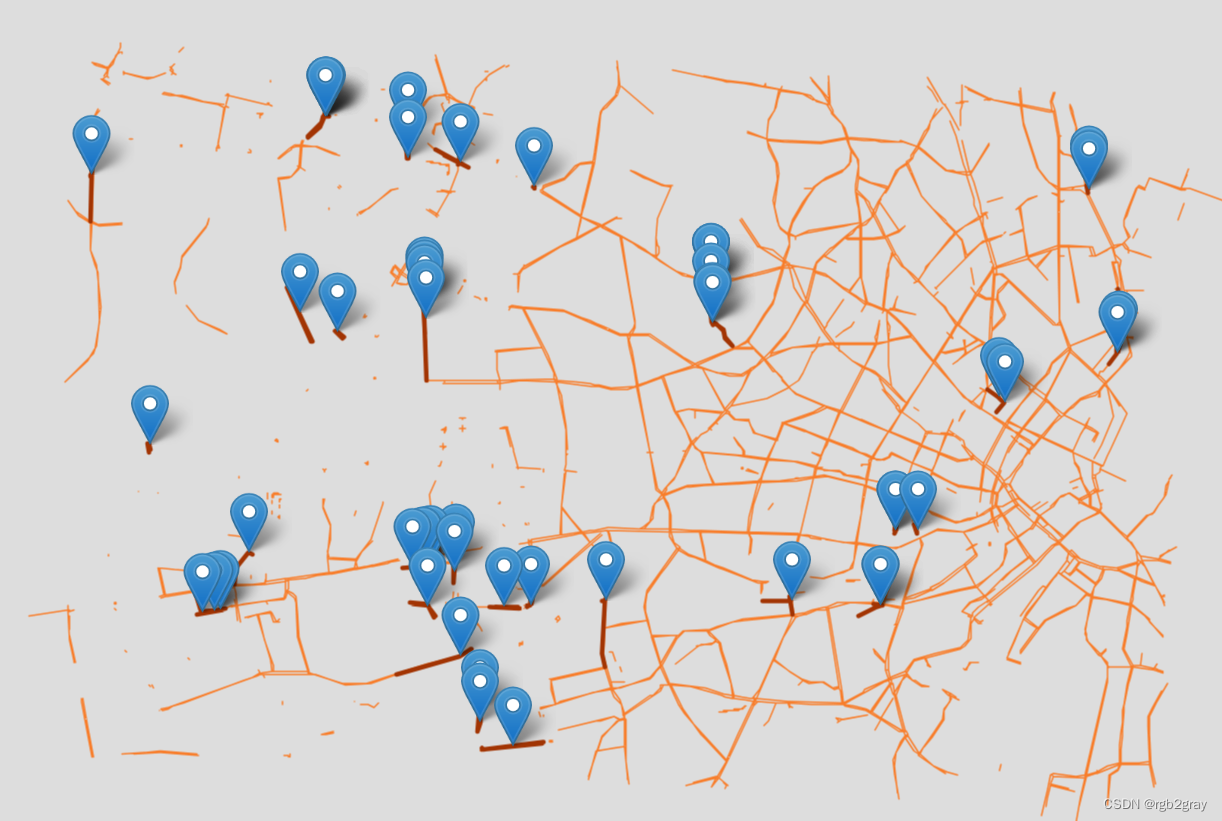
if len(component_gaps) > 0:
print("Interactive map saved at " + ref_results_inter_maps_fp.lstrip("../") + f"component_gaps_{component_min_distance}_reference.html")
else:
print("There are no component gaps to plot.")
Interactive map saved at results/REFERENCE/cph_geodk/maps_interactive/component_gaps_10_reference.html
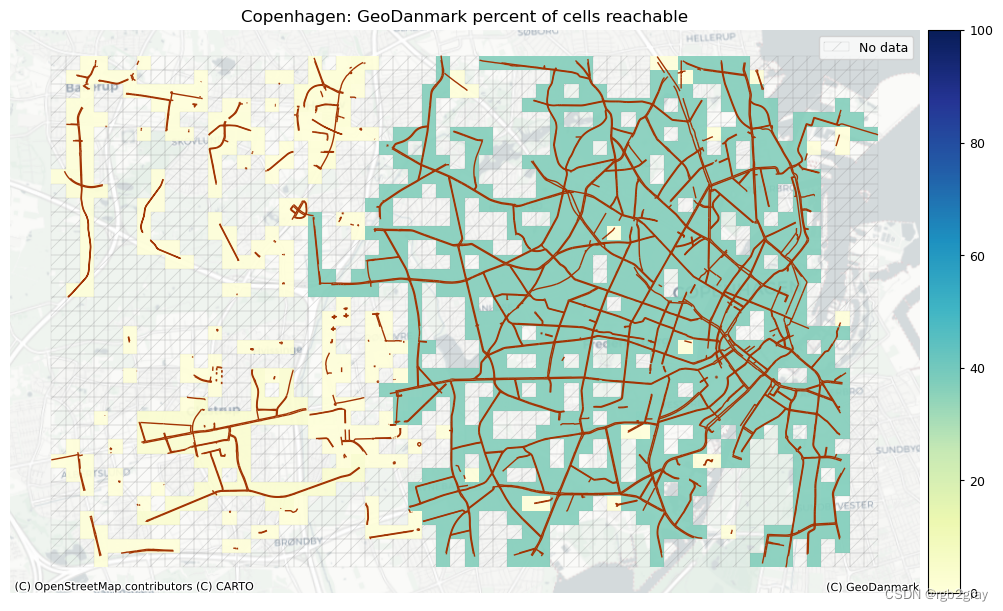
组件连接
在这里,我们可视化每个单元格可以到达的单元格数量之间的差异。 这是对网络连接性的粗略测量,但具有计算成本低的优点,因此能够快速突出网络连接性的明显差异。
ref_components_cell_count = eval_func.count_component_cell_reach(
components_df, grid, "component_ids_ref"
)
grid["cells_reached_ref"] = grid["component_ids_ref"].apply(
lambda x: eval_func.count_cells_reached(x, ref_components_cell_count)
if x != ""
else 0
)
grid["cells_reached_ref_pct"] = grid.apply(
lambda x: np.round((x.cells_reached_ref / len(grid)) * 100, 2), axis=1
)
grid.loc[grid["cells_reached_ref_pct"] == 0, "cells_reached_ref_pct"] = np.NAN
# Plot percent of cells reachable
set_renderer(renderer_map)
fig, ax = plt.subplots(1, 1, figsize=pdict["fsmap"])
# norm for color bars
cbnorm_reach = colors.Normalize(vmin=0, vmax=100)
cbnorm_reach_diff = colors.Normalize(vmin=-100, vmax=100)
from mpl_toolkits.axes_grid1 import make_axes_locatable
divider = make_axes_locatable(ax)
cax = divider.append_axes("right", size="3.5%", pad="1%")
grid.plot(
cax=cax,
ax=ax,
column="cells_reached_ref_pct",
legend=True,
cmap=pdict["seq"],
norm=cbnorm_reach,
alpha=pdict["alpha_grid"],
)
ref_edges_simplified.plot(ax=ax, color=pdict["ref_emp"], linewidth=pdict["line_base"])
# add no data patches
grid[grid["count_ref_edges"].isnull()].plot(
cax=cax,
ax=ax,
facecolor=pdict["nodata_face"],
edgecolor=pdict["nodata_edge"],
linewidth= pdict["line_nodata"],
hatch=pdict["nodata_hatch"],
alpha=pdict["alpha_nodata"],
)
ax.legend(handles=[nodata_patch], loc="upper right")
cx.add_basemap(ax=ax, crs=study_crs, source=cx_tile_2)
cx.add_attribution(ax=ax, text=f"(C) {reference_name}")
txt = ax.texts[-1]
txt.set_position([1,0.00])
txt.set_ha('right')
txt.set_va('bottom')
ax.set_title(area_name+f": {reference_name} percent of cells reachable")
ax.set_axis_off()
plot_func.save_fig(fig,ref_results_static_maps_fp + "percent_cells_reachable_grid_reference")
components_results = {}
components_results["component_count"] = len(ref_components)
components_results["largest_cc_pct_size"] = largest_cc_pct
components_results["largest_cc_length"] = largest_cc_length
components_results["count_component_gaps"] = len(component_gaps)
4.概括
summarize_results = {**density_results, **components_results}
summarize_results["count_dangling_nodes"] = len(dangling_nodes)
summarize_results["count_overshoots"] = len(overshoots)
summarize_results["count_undershoots"] = len(undershoot_nodes)
# Add total node count and total infrastructure length
summarize_results["total_nodes"] = len(ref_nodes_simplified)
summarize_results["total_length"] = ref_edges_simplified.infrastructure_length.sum() / 1000
summarize_results_df = pd.DataFrame.from_dict(summarize_results, orient="index")
summarize_results_df.rename({0: " "}, axis=1, inplace=True)
# Convert length to km
summarize_results_df.loc["largest_cc_length"] = (
summarize_results_df.loc["largest_cc_length"] / 1000
)
summarize_results_df = summarize_results_df.reindex([
'total_length',
'protected_density_m_sqkm',
'unprotected_density_m_sqkm',
'mixed_density_m_sqkm',
'edge_density_m_sqkm',
'total_nodes',
'count_dangling_nodes',
'node_density_count_sqkm',
'dangling_node_density_count_sqkm',
'count_overshoots',
'count_undershoots',
'component_count',
'largest_cc_length',
'largest_cc_pct_size',
'count_component_gaps'
])
rename_metrics = {
"total_length": "Total infrastructure length (km)",
"total_nodes": "Nodes",
"edge_density_m_sqkm": "Bicycle infrastructure density (m/km2)",
"node_density_count_sqkm": "Nodes per km2",
"dangling_node_density_count_sqkm": "Dangling nodes per km2",
"protected_density_m_sqkm": "Protected bicycle infrastructure density (m/km2)",
"unprotected_density_m_sqkm": "Unprotected bicycle infrastructure density (m/km2)",
"mixed_density_m_sqkm": "Mixed protection bicycle infrastructure density (m/km2)",
"component_count": "Components",
"largest_cc_pct_size": "Largest component's share of network length",
"largest_cc_length": "Length of largest component (km)",
"count_component_gaps": "Component gaps",
"count_dangling_nodes": "Dangling nodes",
"count_overshoots": "Overshoots",
"count_undershoots": "Undershoots",
}
summarize_results_df.rename(rename_metrics, inplace=True)
summarize_results_df.style.pipe(format_ref_style)
| Total infrastructure length (km) | 626 |
|---|---|
| Protected bicycle infrastructure density (m/km2) | 2,999 |
| Unprotected bicycle infrastructure density (m/km2) | 455 |
| Mixed protection bicycle infrastructure density (m/km2) | 0 |
| Bicycle infrastructure density (m/km2) | 3,454 |
| Nodes | 4,125 |
| Dangling nodes | 870 |
| Nodes per km2 | 23 |
| Dangling nodes per km2 | 5 |
| Overshoots | 21 |
| Undershoots | 11 |
| Components | 204 |
| Length of largest component (km) | 501 |
| Largest component's share of network length | 80% |
| Component gaps | 52 |
5.保存结果
all_results = {}
all_results["network_density"] = density_results
all_results["count_overshoots"] = len(overshoots)
all_results["count_undershoots"] = len(undershoot_nodes)
all_results["dangling_node_count"] = len(dangling_nodes)
all_results["simplification_outcome"] = simplification_results
all_results["component_analysis"] = components_results
with open(ref_intrinsic_fp, "w") as outfile:
json.dump(all_results, outfile)
# Save summary dataframe
summarize_results_df.to_csv(
ref_results_data_fp + "intrinsic_summary_results.csv", index=True
)
# Save grid with results
with open(ref_intrinsic_grid_fp, "wb") as f:
pickle.dump(grid, f)
from time import strftime
print("Time of analysis: " + strftime("%a, %d %b %Y %H:%M:%S"))
Time of analysis: Mon, 18 Dec 2023 20:21:28