本期图片
❝Doi:https://doi.org/10.1016/j.ccell.2022.02.013
❞
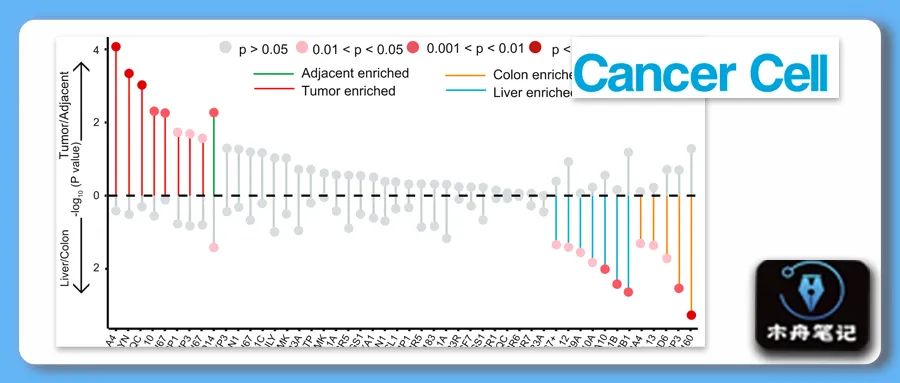
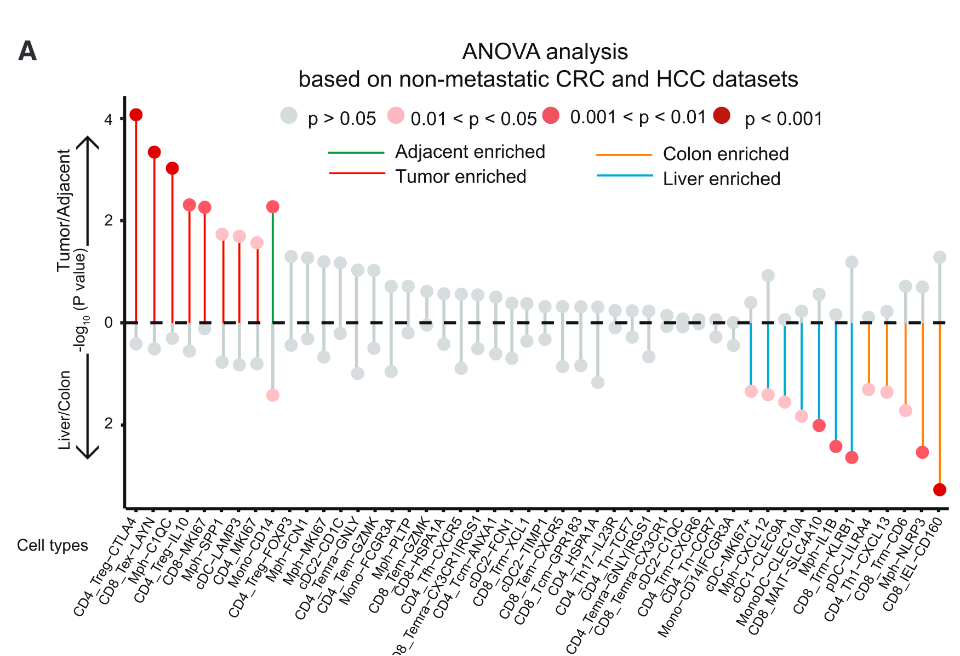
❝Dotplot showing the association of cell populations with different tissue types (organ or tumor). The x axis represents cell types, and the y axis represents the different factors defining tissue types. Cell types in the upper panel are associated with the tissue type (tumor/adjacent normal). Green lines represent cells enriched in adjacent tissue, and red lines represent cells enriched in tumors. Cell types in the bottom panel are associated with if the tissue is liver or colon. Orange lines represent cells enriched in the colon, and blue lines represent cells enriched in the liver. Only the cell types with p < 0.05 were highlighted.
❞
复现结果
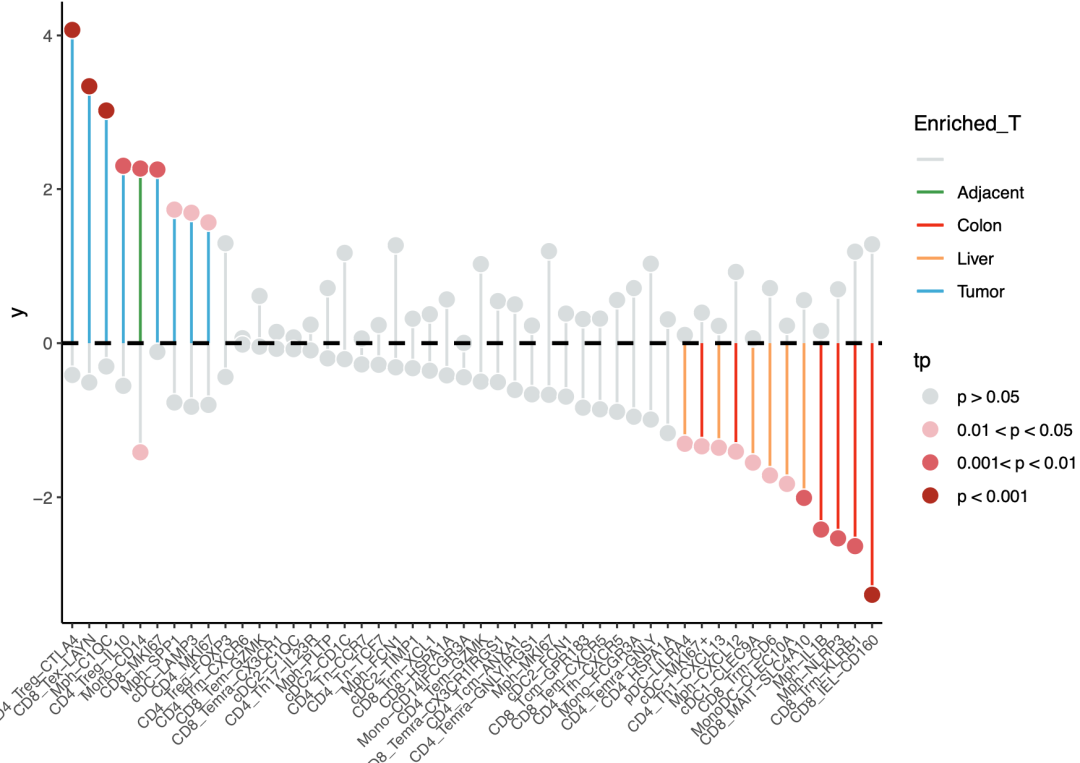
❝有些细节上的代码实在懒得调了,大家见谅。可在AI上微调。使用代码时请按需修改,切勿全部套用。
❞
示例数据和代码领取
木舟笔记永久VIP企划
「权益:」
「木舟笔记所有推文示例数据及代码(「在VIP群里」实时更新」)。
data+code 木舟笔记「科研交流群」。
「收费:」
「169¥/人」。可添加微信:mzbj0002 转账(或扫描下方二维码),或直接在文末打赏。
❝❞
点赞、在看本文,分享至朋友圈集赞30个并保留30分钟,可优惠20¥。
绘图
data <- read.csv("mmc2.csv")
head(data)
df_order <- data[order(data$Tumor_Pvalue),]
df_order <- rbind(
df_order [1:10,],
df_order[11:48,] %>% arrange(desc(Organ_Pvalue))
)
df_order$tp = ifelse(df_order$Tumor_Pvalue < 0.001, "p < 0.001",
ifelse(df_order$Tumor_Pvalue >= 0.001 & df_order$Tumor_Pvalue < 0.01, "0.001< p < 0.01",
ifelse(df_order$Tumor_Pvalue >= 0.01 & df_order$Tumor_Pvalue < 0.05, "0.01 < p < 0.05", "p > 0.05")))
df_order$op = ifelse(df_order$Organ_Pvalue < 0.001, "p < 0.001",
ifelse(df_order$Organ_Pvalue >= 0.001 & df_order$Organ_Pvalue < 0.01, "0.001< p < 0.01",
ifelse(df_order$Organ_Pvalue >= 0.01 & df_order$Organ_Pvalue < 0.05, "0.01 < p < 0.05", "p > 0.05")))
df_order$tp <-factor(df_order$tp,levels = c("p > 0.05","0.01 < p < 0.05","0.001< p < 0.01","p < 0.001"))
df_order$op <- factor(df_order$op,levels = c("p > 0.05","0.01 < p < 0.05","0.001< p < 0.01","p < 0.001"))
df_order$Enriched <- factor(df_order$Enriched, levels = c('Tumor','Adjacent','Liver','Colon') )
df_order$Celltype <- factor(df_order$Celltype, levels = df_order$Celltype)
head(df_order)
## plot
ggplot(df_order)+
geom_segment(aes(x = Celltype,xend = Celltype,
y = 0,yend = -log10(Tumor_Pvalue),
color = Enriched_T),size = 0.7) +
geom_segment(aes(x = Celltype,xend = Celltype,
y = 0,yend = log10(Organ_Pvalue),
color = Enriched_O),size = 0.7) +
scale_color_manual(values = c('#d8ddde','#3f9c49','#ed301a','#faa25c','#3fabd6'))+
geom_point(
aes(x=Celltype, y=-log10(Tumor_Pvalue),fill = tp),
size = 4,shape = 21,color = 'white'
) +
scale_fill_manual(values = c('#d8ddde','#f1bbc0','#db5e64','#b12c20')) +
geom_point(
aes(x=Celltype, y=log10(Organ_Pvalue),fill = op),
size = 4,shape = 21,color = 'white'
) +
geom_hline(yintercept = 0, linetype = "dashed",size = 1) +
theme_classic()+
xlab('')+
theme(axis.text.x = element_text(angle = 45, hjust = 1,size = 8))
ggsave('dotplot.pdf',width = 8,height = 6)往期内容
资源汇总 | 2022 木舟笔记原创推文合集(附数据及代码领取方式)
CNS图表复现|生信分析|R绘图 资源分享&讨论群!
R绘图 | 浅谈散点图及其变体的作图逻辑
这图怎么画| 有点复杂的散点图
这图怎么画 | 相关分析棒棒糖图
组学生信| Front Immunol |基于血清蛋白质组早期诊断标志筛选的简单套路
(免费教程+代码领取)|跟着Cell学作图系列合集
Q&A | 如何在论文中画出漂亮的插图?
跟着 Cell 学作图 | 桑葚图(ggalluvial)
R实战 | Lasso回归模型建立及变量筛选
跟着 NC 学作图 | 互作网络图进阶(蛋白+富集通路)(Cytoscape)
R实战 | 给聚类加个圈圈(ggunchull)
R实战 | NGS数据时间序列分析(maSigPro)
跟着 Cell 学作图 | 韦恩图(ggVennDiagram)






![[项目管理-33/创业之路-87/管理者与领导者-127]:如何提升自己项目管理的能力和水平](https://img-blog.csdnimg.cn/6a57e0c070d94ecc9b0ba63959c35c78.png)