我好像做了一个愚蠢的测试
rm(list=ls())
suppressPackageStartupMessages({
library(SingleCellExperiment)
library(scMerge)
library(scater)
library(Matrix)
})
setwd("/Users/yxk/Desktop/test/R_parallel/")
load("./data/exprsMat.RData")
load("./data/clust.RData")
load("./data/pseudobulk_sample_list.RData")
load("./data/pseudobulk_sample.RData")
load("./data/use_bpparm.RData")
load("./data/res.RData")
#' @importFrom ruv replicate.matrix
#' @importFrom methods as is
aggregate.Matrix <- function(x, groupings=NULL) {
if (!methods::is(x,'Matrix')) {
x <- methods::as(as.matrix(x), "CsparseMatrix")
}
groupings2 <- paste("A", groupings, sep = "")
if (length(unique(groupings2)) > 1) {
mapping <- methods::as(ruv::replicate.matrix(groupings2), "CsparseMatrix")
colnames(mapping) <- substring(colnames(mapping), 2)
mapping <- mapping[, levels(factor(groupings))]
} else {
mapping <- methods::as(matrix(rep(1, length(groupings2)), ncol = 1), "CsparseMatrix")
colnames(mapping) <- unique(groupings)
}
result <- t(mapping) %*% x
return(result)
}
create_pseudoBulk_parallel = function (exprsMat, cell_info, k_fold = 30, use_bpparam = BiocParallel::SerialParam())
{
#browser()
k_fold <- min(ncol(exprsMat), k_fold)
cv <- cvTools::cvFolds(ncol(exprsMat), K = k_fold)
exprsMat_pseudo <- BiocParallel::bplapply(seq_len(k_fold),
function(i) {
subset_idx <- cv$subsets[cv$which == i]
cellType_tab <- table(droplevels(factor(cell_info[subset_idx])))
cellTypes_n_mat <- matrix(rep(cellType_tab, nrow(exprsMat)),
nrow = length(cellType_tab), byrow = FALSE)
rownames(cellTypes_n_mat) <- names(cellType_tab)
res <- aggregate.Matrix(t(exprsMat[, subset_idx]),
cell_info[subset_idx])
cellTypes_n_mat <- cellTypes_n_mat[rownames(res),
]
res <- res/cellTypes_n_mat
rownames(res) <- paste(rownames(res), i, sep = "_")
res
}, BPPARAM = use_bpparam)
exprsMat_pseudo <- do.call(rbind, exprsMat_pseudo)
return(exprsMat_pseudo)
}
create_pseudoBulk_no = function (exprsMat, cell_info, k_fold = 30)
{
#browser()
k_fold <- min(ncol(exprsMat), k_fold)
cv <- cvTools::cvFolds(ncol(exprsMat), K = k_fold)
exprsMat_pseudo =list()
for (i in seq_len(k_fold)){
subset_idx <- cv$subsets[cv$which == i]
cellType_tab <- table(droplevels(factor(cell_info[subset_idx])))
cellTypes_n_mat <- matrix(rep(cellType_tab, nrow(exprsMat)),
nrow = length(cellType_tab), byrow = FALSE)
rownames(cellTypes_n_mat) <- names(cellType_tab)
res <- aggregate.Matrix(t(exprsMat[, subset_idx]),
cell_info[subset_idx])
cellTypes_n_mat <- cellTypes_n_mat[rownames(res),
]
res <- res/cellTypes_n_mat
rownames(res) <- paste(rownames(res), i, sep = "_")
exprsMat_pseudo[[i]] = res
}
exprsMat_pseudo <- do.call(rbind, exprsMat_pseudo)
return(exprsMat_pseudo)
}
set.seed(1)
i =1
res1 <- create_pseudoBulk_parallel(exprsMat[, pseudobulk_sample ==
pseudobulk_sample_list[i]], clust[[i]], k_fold = 30,
use_bpparam = use_bpparam)
set.seed(1)
i =1
res2 <- create_pseudoBulk_no(exprsMat[, pseudobulk_sample ==
pseudobulk_sample_list[i]],
clust[[i]], k_fold = 30
)
print("done")
# for (i in seq_along(pseudobulk_sample_list)) {
# res <- create_pseudoBulk_parallel(exprsMat[, pseudobulk_sample ==
# pseudobulk_sample_list[i]], clust[[i]], k_fold = 30,
# use_bpparam = use_bpparam)
# }
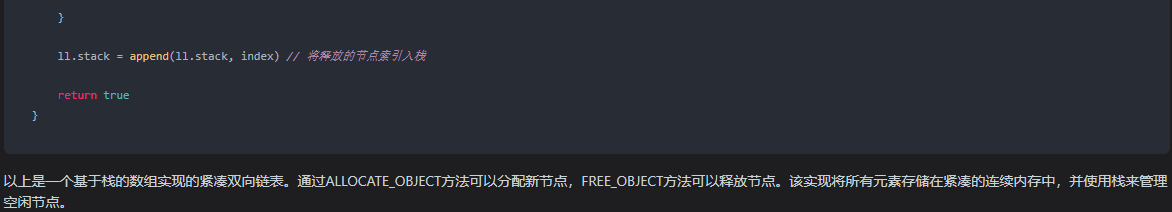
首先注意一个随机种子的问题,否则这个结果就会不一样