# https://mne.tools/stable/auto_tutorials/forward/35_eeg_no_mri.html
# 本教程说明如何使用标准模板 MRI 主题根据 EEG 数据计算前向算子
#
# 成人模板 MRI (fsaverage)
# 首先我们展示如何fsaverage用作替代subject
# Authors: Alexandre Gramfort <alexandre.gramfort@inria.fr>
# Joan Massich <mailsik@gmail.com>
# Eric Larson <larson.eric.d@gmail.com>
#
# License: BSD-3-Clause
import os.path as op
import numpy as np
import mne
from mne.datasets import eegbci
from mne.datasets import fetch_fsaverage
# Download fsaverage files
fs_dir = fetch_fsaverage(verbose=True)
subjects_dir = op.dirname(fs_dir)
# The files live in:
subject = "fsaverage"
trans = "fsaverage" # MNE has a built-in fsaverage transformation
src = op.join(fs_dir, "bem", "fsaverage-ico-5-src.fif")
bem = op.join(fs_dir, "bem", "fsaverage-5120-5120-5120-bem-sol.fif")
#加载数据 BCI的
# (raw_fname,) = eegbci.load_data(subject=1, runs=[6])
# raw = mne.io.read_raw_edf(raw_fname, preload=True)
raw = mne.io.read_raw_eeglab('eeg_data/sub2.set')
# Clean channel names to be able to use a standard 1005 montage
# new_names = dict(
# (ch_name, ch_name.rstrip(".").upper().replace("Z", "z").replace("FP", "Fp"))
# for ch_name in raw.ch_names
# )
# raw.rename_channels(new_names)
# Read and set the EEG electrode locations, which are already in fsaverage's
# space (MNI space) for standard_1020:
# montage = mne.channels.make_standard_montage("standard_1005")
# raw.set_montage(montage)
raw.set_eeg_reference(projection=True) # needed for inverse modeling
# Check that the locations of EEG electrodes is correct with respect to MRI
# mne.viz.plot_alignment(
# raw.info,
# src=src,
# eeg=["original", "projected"],
# trans=trans,
# show_axes=True,
# mri_fiducials=True,
# dig="fiducials",
# )
#Setup source space and compute forward
fwd = mne.make_forward_solution(
raw.info, trans=trans, src=src, bem=bem, eeg=True, mindist=5.0, n_jobs=None
)
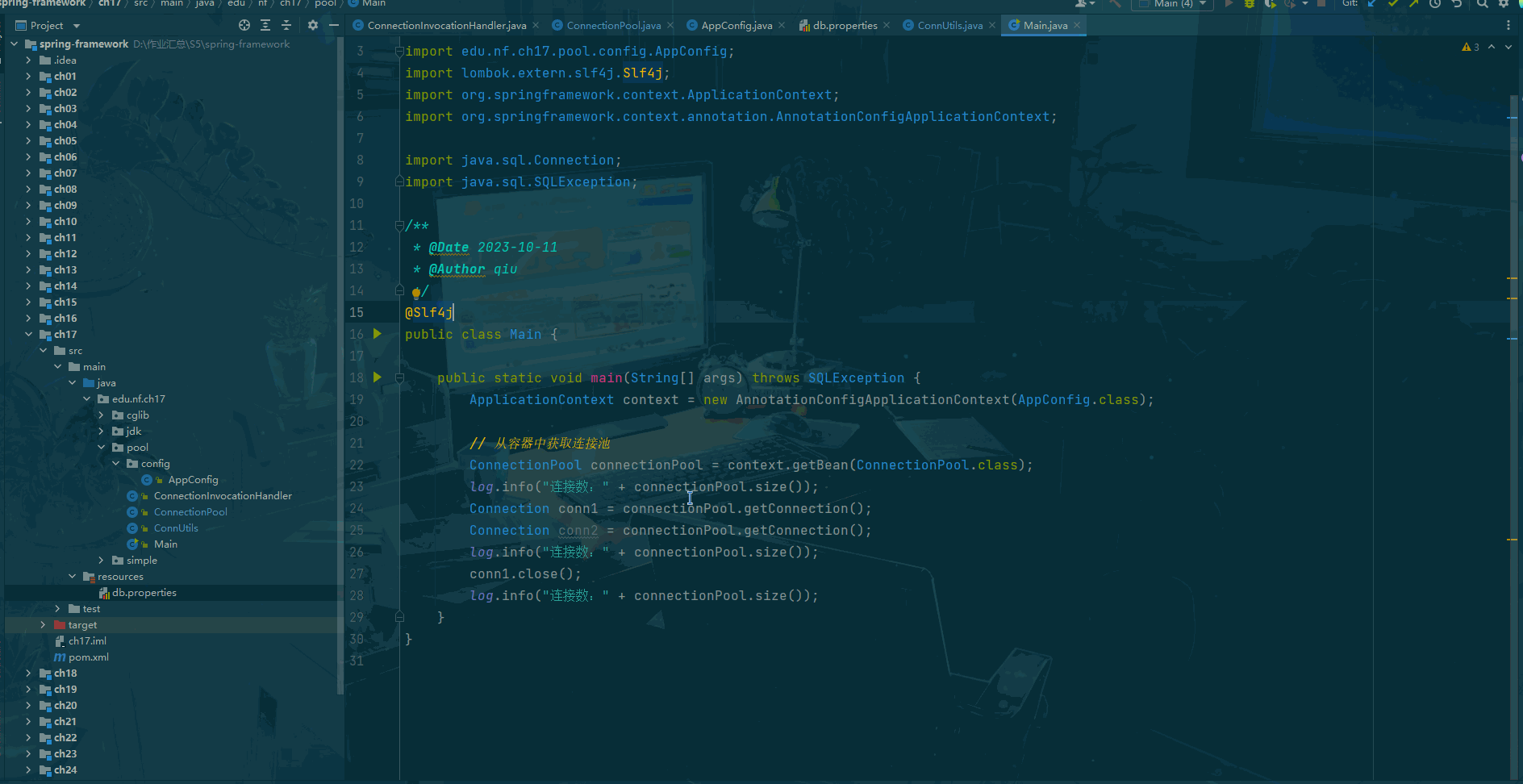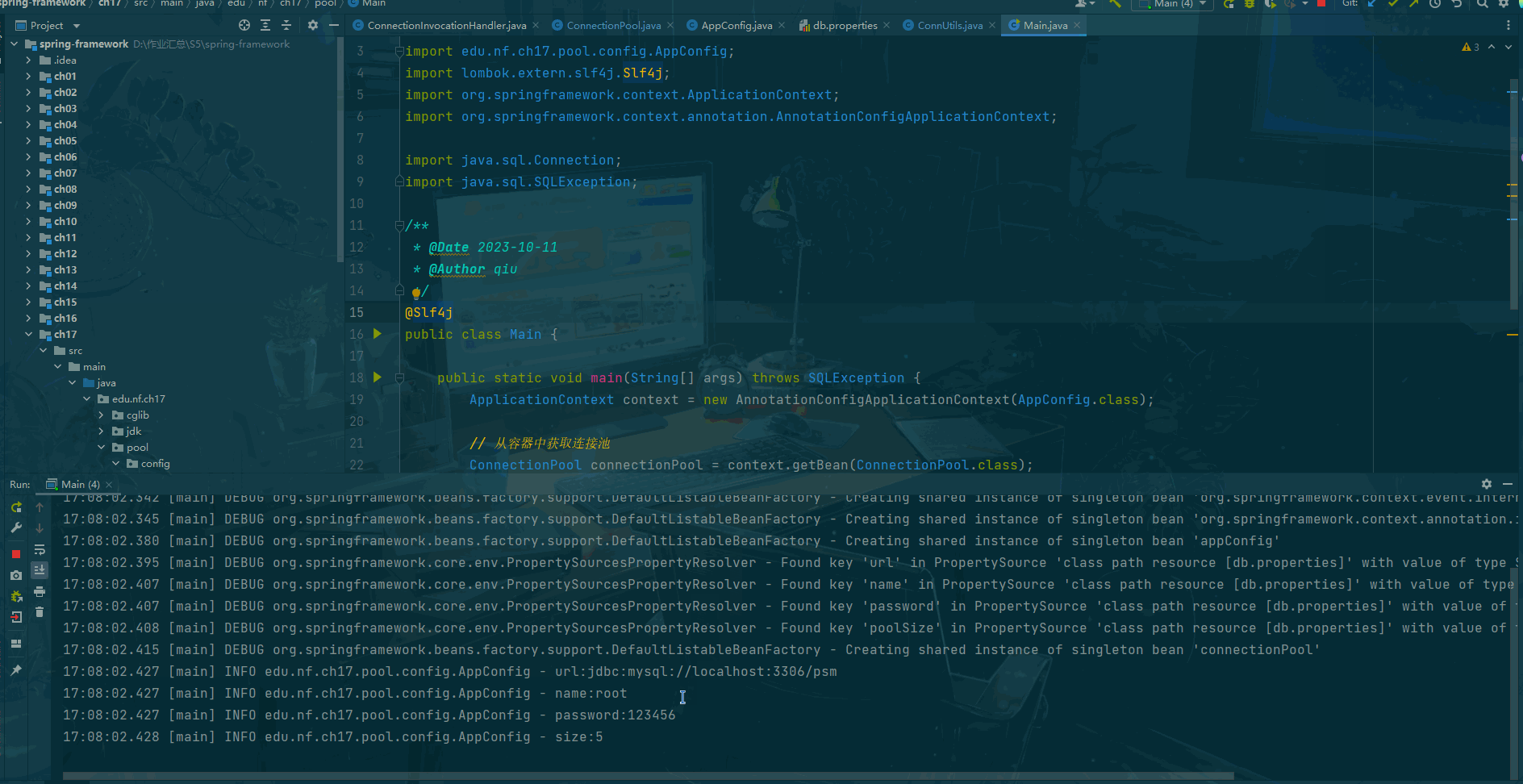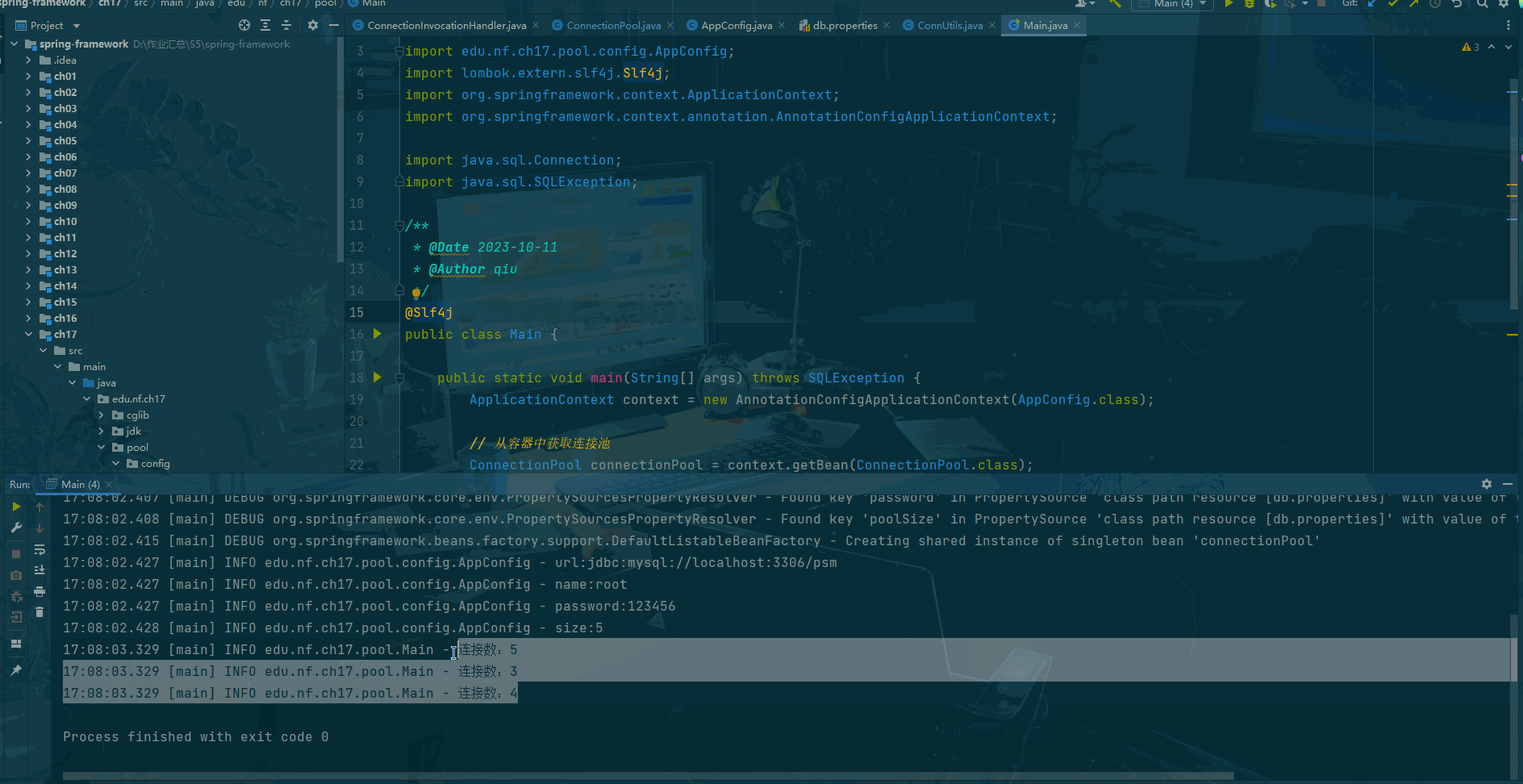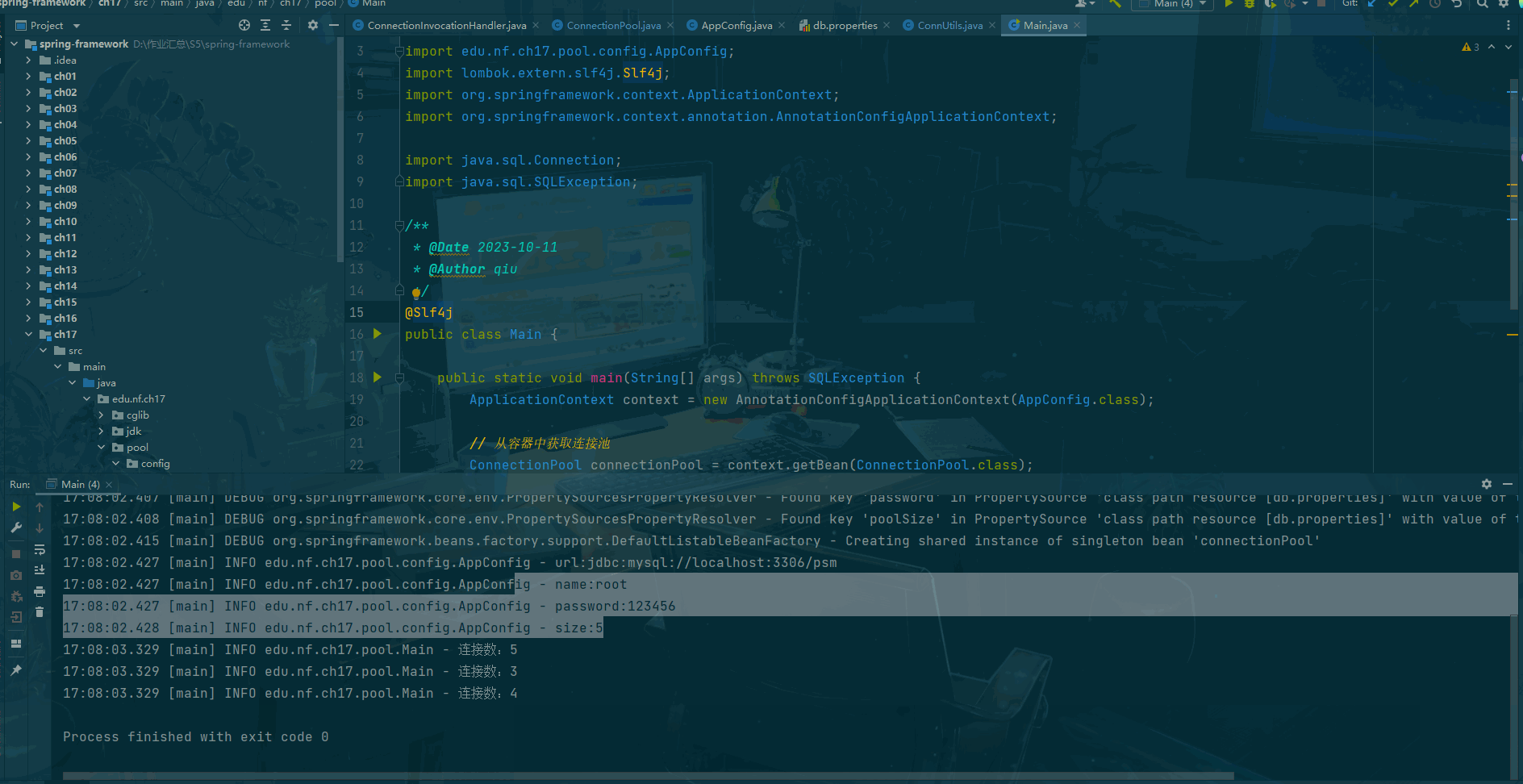
print()
#保存前向解到文件中
# mne.write_forward_solution('solution_bci4.fif',fwd,overwrite=True)
# 保存出问题,暂时先不保存







![[linux] 把txt文本文件分成10个子文件,并保存。 linux命令](https://img-blog.csdnimg.cn/5f4e917ba5be43388a08e938f7b7a7fa.png)

![2023年全球及中国环球影城主题公园数量、游客人数及收入分析[图]](https://img-blog.csdnimg.cn/img_convert/83cdb6784d1520552a73d23535e7264c.png)