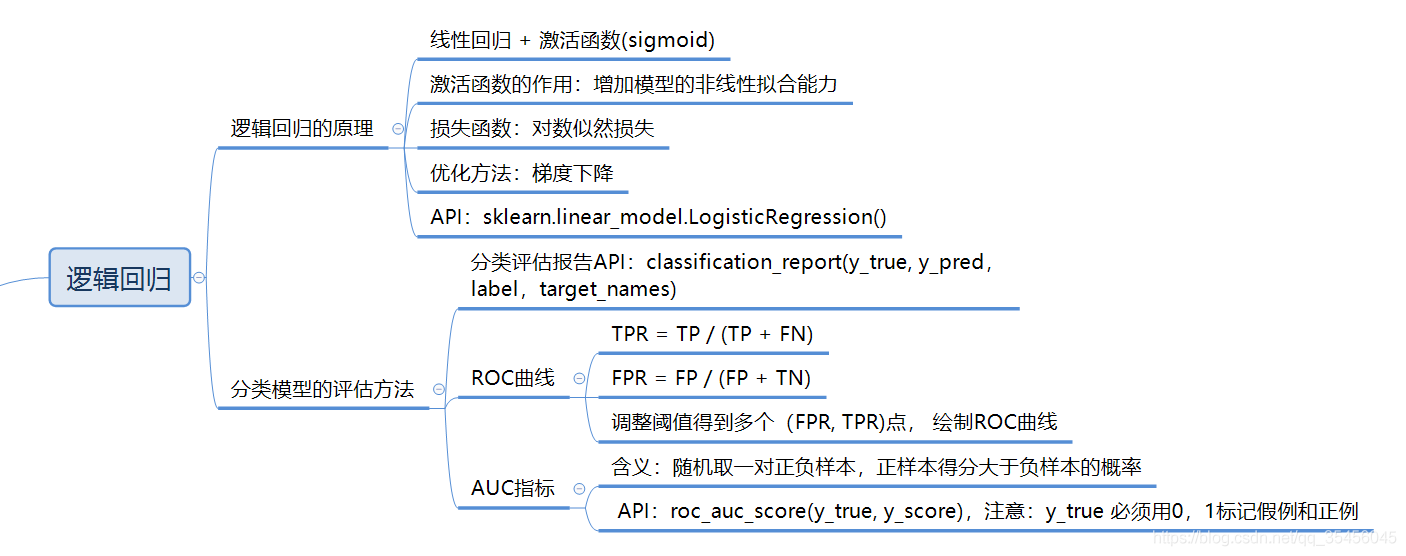
导入包
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report,roc_auc_score
读取数据集
# 特征名称
feature_names = ['Sample code number', 'Clump Thickness', 'Uniformity of Cell Size', 'Uniformity of Cell Shape',
'Marginal Adhesion', 'Single Epithelial Cell Size', 'Bare Nuclei', 'Bland Chromatin',
'Normal Nucleoli', 'Mitoses', 'Class']
data = pd.read_csv(
"https://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.data",
names=feature_names)
data.head()
| Sample code number | Clump Thickness | Uniformity of Cell Size | Uniformity of Cell Shape | Marginal Adhesion | Single Epithelial Cell Size | Bare Nuclei | Bland Chromatin | Normal Nucleoli | Mitoses | Class | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1000025 | 5 | 1 | 1 | 1 | 2 | 1 | 3 | 1 | 1 | 2 |
| 1 | 1002945 | 5 | 4 | 4 | 5 | 7 | 10 | 3 | 2 | 1 | 2 |
| 2 | 1015425 | 3 | 1 | 1 | 1 | 2 | 2 | 3 | 1 | 1 | 2 |
| 3 | 1016277 | 6 | 8 | 8 | 1 | 3 | 4 | 3 | 7 | 1 | 2 |
| 4 | 1017023 | 4 | 1 | 1 | 3 | 2 | 1 | 3 | 1 | 1 | 2 |
列名解释
在实际工作中,要弄清楚每一个肿瘤特征代表什么含义,这样才能做好异常值缺失值的处理
- Sample code number 样本编号 用不到
- Clump Thickness 肿瘤特征1
- Uniformity of Cell Size 肿瘤特征2
- Uniformity of Cell Shape 肿瘤特征3
- Marginal Adhesion 肿瘤特征4
- Single Epithelial Cell Size 肿瘤特征5
- Bare Nuclei 肿瘤特征6
- Bland Chromatin 肿瘤特征7
- Normal Nucleoli 肿瘤特征8
- Mitoses 肿瘤特征9
- Class 肿瘤的种类
查看目标值
- 2表示良性,444条数据
- 4表示恶性,239条数据
data.Class.value_counts()
2 458
4 241
Name: Class, dtype: int64
数据基本处理 缺失值处理
# 替换缺失值
data = data.replace(to_replace='?', value=np.nan)
# 删除缺失值的样本
data = data.dropna()
划分数据集
X = data[feature_names[0:-1]] # ==> X = data.iloc[:, 0:-1]
y = data["Class"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
特征工程 标准化
transform = StandardScaler()
X_train = transform.fit_transform(X_train)
X_test = transform.fit_transform(X_test)
模型训练
lr_model = LogisticRegression()
lr_model.fit(X_train, y_train)
LogisticRegression()
模型评估(准确率、精确率、召回率)
模型在训练集上的准确率
lr_model.score(X_train, y_train)
0.9688644688644689
模型在测试集上的准确率
lr_model.score(X_test, y_test)
0.9781021897810219
预测精确率、召回率
y_pred = lr_model.predict(X_test)
report = classification_report(y_test, y_pred, labels=(2, 4), target_names=("良性", "恶性"))
print(report)
precision recall f1-score support
良性 0.99 0.98 0.98 96
恶性 0.95 0.98 0.96 41
accuracy 0.98 137
macro avg 0.97 0.98 0.97 137
weighted avg 0.98 0.98 0.98 137






![java八股文面试[java基础]——CGLIB动态代理与JDK动态代理](https://img-blog.csdnimg.cn/ee9bd2c2eee24507996f63a51ee1bcea.png)
