https://scikit-learn.org/stable/modules/preprocessing.html
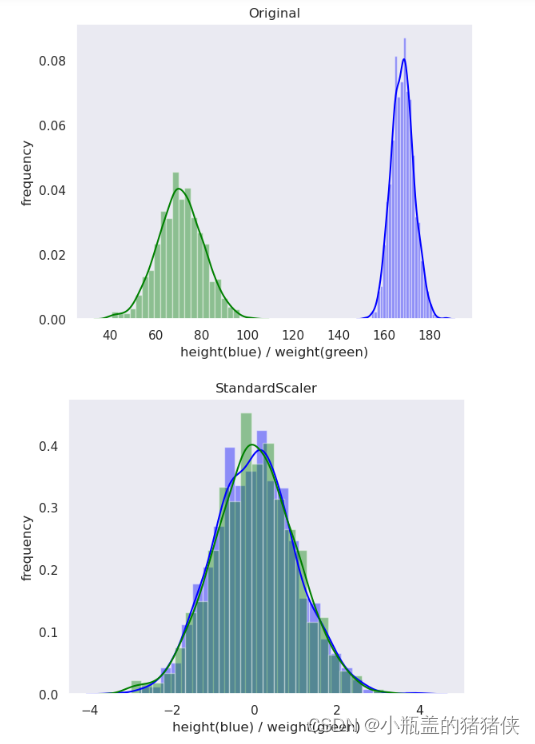
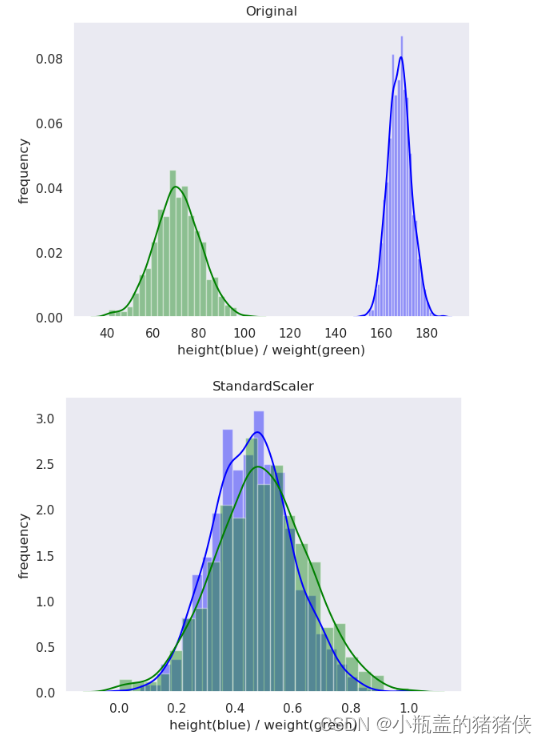
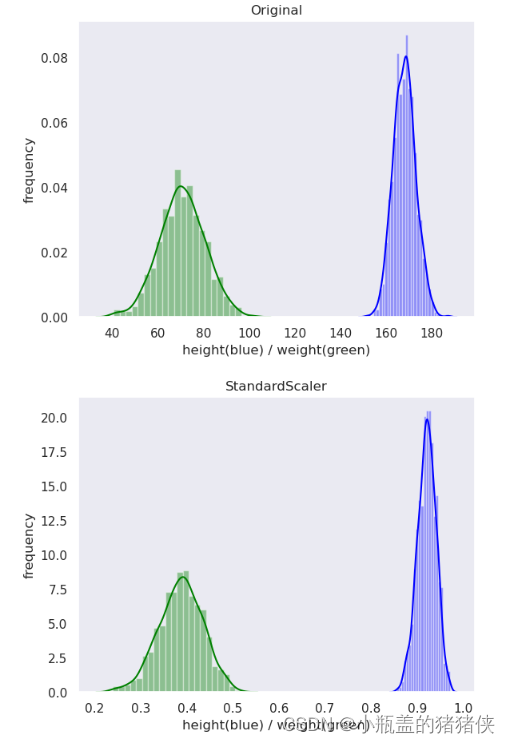
标准化
公式 : 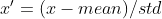
优点:适用大多数类型的数据,标准化之后的数据是以0为均值,方差为1的正态分布
缺点 :是一种中心化方法,会改变原有数据得分布结构
转换区间:均值为0,方差为1的标准正态分布
适用场景:不适合用于稀疏数据的处理
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import preprocessing
def plot(data, title):
sns.set_style('dark')
f, ax = plt.subplots()
ax.set(ylabel='frequency')
ax.set(xlabel='height(blue) / weight(green)')
ax.set(title=title)
sns.distplot(data[:, 0:1], color='blue')
sns.distplot(data[:, 1:2], color='green')
plt.savefig(title + '.png')
plt.show()
np.random.seed(42)
height = np.random.normal(loc=168, scale=5, size=1000).reshape(-1, 1)
weight = np.random.normal(loc=70, scale=10, size=1000).reshape(-1, 1)
original_data = np.concatenate((height, weight), axis=1)
plot(original_data, 'Original')
standard_scaler_data = preprocessing.StandardScaler().fit_transform(original_data)
plot(standard_scaler_data, 'StandardScaler')
最大最小归一化
公式 :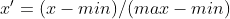
优点:应用广泛,能较好的保持原有数据分布结构
缺点 :.对异常值(离群值)的存在非常敏感
转换区间:[0,1]
适用场景:不适合用于稀疏数据的处理
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import preprocessing
def plot(data, title):
sns.set_style('dark')
f, ax = plt.subplots()
ax.set(ylabel='frequency')
ax.set(xlabel='height(blue) / weight(green)')
ax.set(title=title)
sns.distplot(data[:, 0:1], color='blue')
sns.distplot(data[:, 1:2], color='green')
plt.savefig(title + '.png')
plt.show()
np.random.seed(42)
height = np.random.normal(loc=168, scale=5, size=1000).reshape(-1, 1)
weight = np.random.normal(loc=70, scale=10, size=1000).reshape(-1, 1)
original_data = np.concatenate((height, weight), axis=1)
plot(original_data, 'Original')
standard_scaler_data = preprocessing.MinMaxScaler().fit_transform(original_data)
plot(standard_scaler_data, 'StandardScaler')
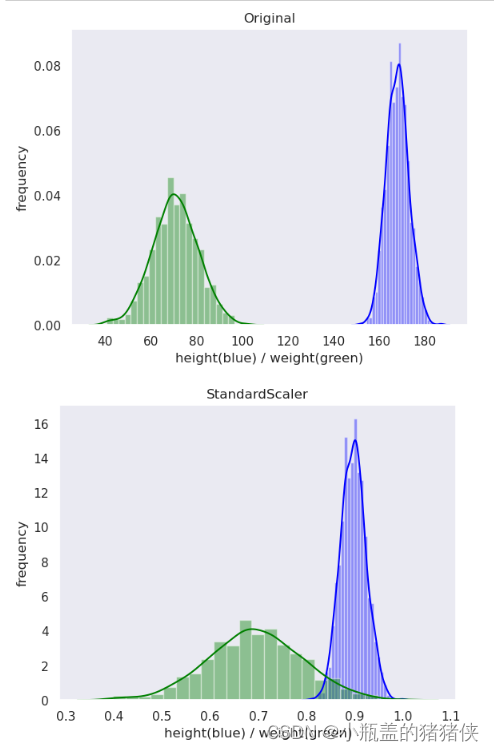
MaxAbs归一化
公式 :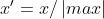
优点:保持原有数据分布结构
缺点 :.对异常值(离群值)的存在非常敏感
转换区间:[-1,1]
适用场景:稀疏数据、稀疏CSR或CSC矩阵
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import preprocessing
def plot(data, title):
sns.set_style('dark')
f, ax = plt.subplots()
ax.set(ylabel='frequency')
ax.set(xlabel='height(blue) / weight(green)')
ax.set(title=title)
sns.distplot(data[:, 0:1], color='blue')
sns.distplot(data[:, 1:2], color='green')
plt.savefig(title + '.png')
plt.show()
np.random.seed(42)
height = np.random.normal(loc=168, scale=5, size=1000).reshape(-1, 1)
weight = np.random.normal(loc=70, scale=10, size=1000).reshape(-1, 1)
original_data = np.concatenate((height, weight), axis=1)
plot(original_data, 'Original')
standard_scaler_data = preprocessing.MinMaxScaler().fit_transform(original_data)
plot(standard_scaler_data, 'StandardScaler')
正则化
公式 :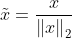 其中
其中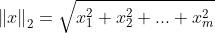
优点:单向量上来实现这正则化的功能
缺点 :.
转换区间:
适用场景:经常被使用在分类与聚类中。
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import preprocessing
def plot(data, title):
sns.set_style('dark')
f, ax = plt.subplots()
ax.set(ylabel='frequency')
ax.set(xlabel='height(blue) / weight(green)')
ax.set(title=title)
sns.distplot(data[:, 0:1], color='blue')
sns.distplot(data[:, 1:2], color='green')
plt.savefig(title + '.png')
plt.show()
np.random.seed(42)
height = np.random.normal(loc=168, scale=5, size=1000).reshape(-1, 1)
weight = np.random.normal(loc=70, scale=10, size=1000).reshape(-1, 1)
original_data = np.concatenate((height, weight), axis=1)
plot(original_data, 'Original')
standard_scaler_data = preprocessing.Normalizer().fit_transform(original_data)
plot(standard_scaler_data, 'StandardScaler')
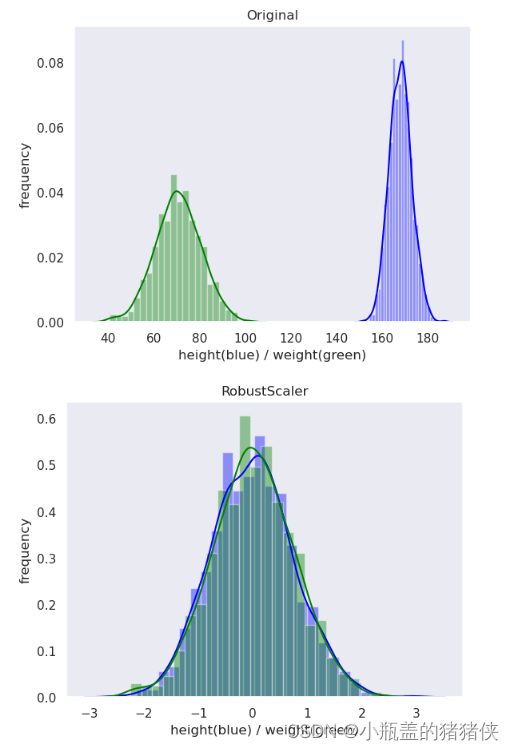
Robust归一化
公式 :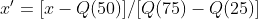 Q代表分位数
Q代表分位数
优点:能最大限度地保留数据集中的异常(离群点)
缺点 :.
转换区间:
适用场景:最大限度保留数据集中的异常(离群值)
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import preprocessing
def plot(data, title):
sns.set_style('dark')
f, ax = plt.subplots()
ax.set(ylabel='frequency')
ax.set(xlabel='height(blue) / weight(green)')
ax.set(title=title)
sns.distplot(data[:, 0:1], color='blue')
sns.distplot(data[:, 1:2], color='green')
plt.savefig(title + '.png')
plt.show()
np.random.seed(42)
height = np.random.normal(loc=168, scale=5, size=1000).reshape(-1, 1)
weight = np.random.normal(loc=70, scale=10, size=1000).reshape(-1, 1)
original_data = np.concatenate((height, weight), axis=1)
plot(original_data, 'Original')
standard_scaler_data = preprocessing.RobustScaler().fit_transform(original_data)
plot(standard_scaler_data, 'RobustScaler')