1写在前面
不知道各位最近过得怎么样,昨天去修了脚🦶,感觉自己马上就要迈入油腻中年人的行列了。🥲
不过说实话,还是挺舒服的,值得再去一次。😅
接着更一下WGCNA的教程吧,还是值得大家好好学习一下的。🙃
今天介绍的是分步法创建网络与识别模块,这种比较适合有个性化需求的人使用,如果你是懒癌晚期,可以直接看上一篇教程。😘
2用到的包
rm(list = ls())
library(tidyverse)
library(WGCNA)
3示例数据
我们还是和之前一样,把之前清洗好的输入数据拿出来吧。😗
load("./Consensus-dataInput.RData")
4提取数据集个数
首先和之前一样提取一下我们的数据集个数,后面会用到。🤓
nSets <- checkSets(multiExpr)$nSets
5挑选软阈值并可视化
5.1 创建power
powers <- c(seq(4,10,by=1), seq(12,20, by=2))
5.2 计算power
和之前一样,我们为每个数据集计算一下power,挑选软阈值。🥰
powerTables = vector(mode = "list", length = nSets)
for (set in 1:nSets){
powerTables[[set]] = list(data = pickSoftThreshold(multiExpr[[set]]$data, powerVector=powers,
verbose = 2)[[2]])
collectGarbage()
}
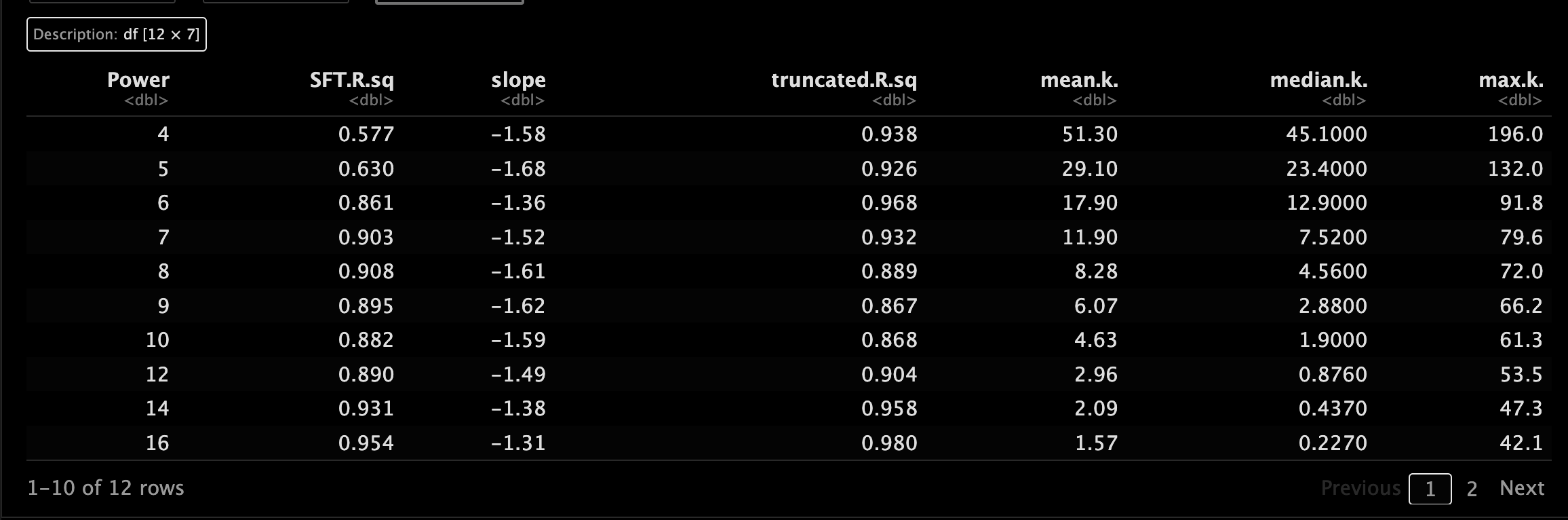
5.3 可视化相关参数设置
设置一下绘图的参数,方便可视化。😂
colors = c("black", "red")
plotCols = c(2,5,6,7)
colNames = c("Scale Free Topology Model Fit", "Mean connectivity", "Median connectivity",
"Max connectivity")
ylim = matrix(NA, nrow = 2, ncol = 4);
for (set in 1:nSets)
{
for (col in 1:length(plotCols))
{
ylim[1, col] = min(ylim[1, col], powerTables[[set]]$data[, plotCols[col]], na.rm = TRUE);
ylim[2, col] = max(ylim[2, col], powerTables[[set]]$data[, plotCols[col]], na.rm = TRUE);
}
}
5.4 可视化一下吧
来绘一下图吧。🐵
sizeGrWindow(8, 6)
par(mfcol = c(2,2));
par(mar = c(4.2, 4.2 , 2.2, 0.5))
cex1 = 0.7;
for (col in 1:length(plotCols)) for (set in 1:nSets)
{
if (set==1)
{
plot(powerTables[[set]]$data[,1], -sign(powerTables[[set]]$data[,3])*powerTables[[set]]$data[,2],
xlab="Soft Threshold (power)",ylab=colNames[col],type="n", ylim = ylim[, col],
main = colNames[col]);
addGrid();
}
if (col==1)
{
text(powerTables[[set]]$data[,1], -sign(powerTables[[set]]$data[,3])*powerTables[[set]]$data[,2],
labels=powers,cex=cex1,col=colors[set]);
} else
text(powerTables[[set]]$data[,1], powerTables[[set]]$data[,plotCols[col]],
labels=powers,cex=cex1,col=colors[set]);
if (col==1)
{
legend("bottomright", legend = setLabels, col = colors, pch = 20) ;
} else
legend("topright", legend = setLabels, col = colors, pch = 20) ;
}
6网络邻接的计算
网络构建首先需要使用软阈值计算各个数据集中的邻接关系,前面计算的power是6哦。😂
softPower <- 6
adjacencies <- array(0, dim = c(nSets, nGenes, nGenes))
for (set in 1:nSets){
adjacencies[set, , ] <- abs(cor(multiExpr[[set]]$data, use = "p"))^softPower
}
7拓扑重叠的计算
接着我们把邻接矩阵转换成拓扑重叠矩阵,也就是计算一下我们常说的TOM。🐵
TOM <- array(0, dim = c(nSets, nGenes, nGenes))
for (set in 1:nSets){
TOM[set, , ] <- TOMsimilarity(adjacencies[set, , ])
}

8给TOM做个scale
8.1 scale一下
不同数据集的TOM可能具有不同的统计特性,可能会导致偏差。😭
为了让两个数据集有可比性,我们需要做一下scale。😋
scaleP = 0.95
set.seed(12345)
nSamples = as.integer(1/(1-scaleP) * 1000)
scaleSample = sample(nGenes*(nGenes-1)/2, size = nSamples)
TOMScalingSamples = list()
scaleQuant = rep(1, nSets)
scalePowers = rep(1, nSets)
for (set in 1:nSets){
TOMScalingSamples[[set]] = as.dist(TOM[set, , ])[scaleSample]
scaleQuant[set] = quantile(TOMScalingSamples[[set]],
probs = scaleP, type = 8)
if (set>1){
scalePowers[set] = log(scaleQuant[1])/log(scaleQuant[set])
TOM[set, ,] = TOM[set, ,]^scalePowers[set]
}
}
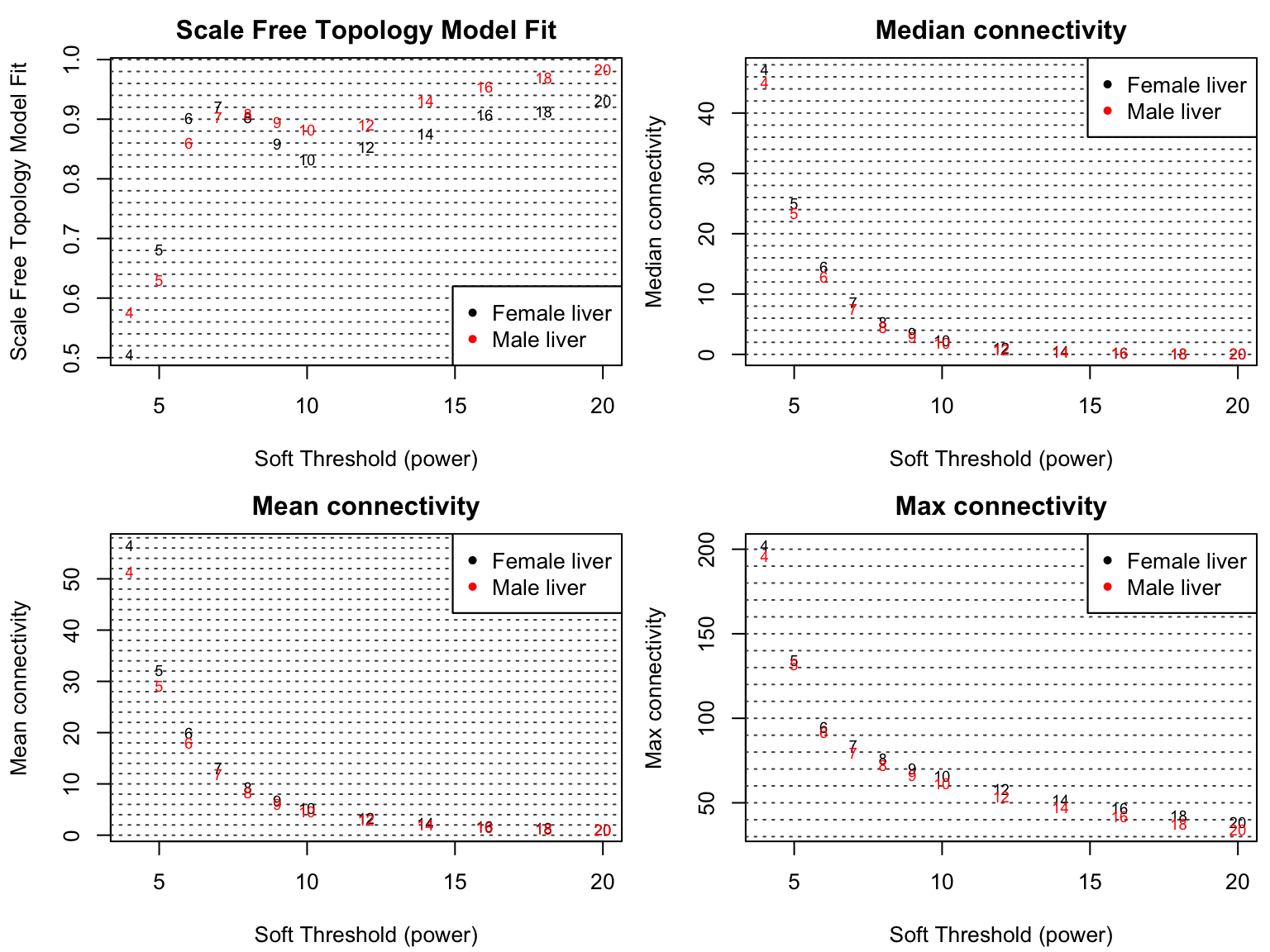
8.2 可视化scale效果
我们做个Q-Q plot来看下scale的效果吧。🤨
做完以后离参考线更近了,当然这种变化比较小就是了。🤣
scaledTOMSamples = list()
for (set in 1:nSets){
scaledTOMSamples[[set]] = TOMScalingSamples[[set]]^scalePowers[set]
}
sizeGrWindow(6,6)
qqUnscaled = qqplot(TOMScalingSamples[[1]], TOMScalingSamples[[2]], plot.it = T, cex = 0.6,
xlab = paste("TOM in", setLabels[1]), ylab = paste("TOM in", setLabels[2]),
main = "Q-Q plot of TOM", pch = 20)
qqScaled = qqplot(scaledTOMSamples[[1]], scaledTOMSamples[[2]], plot.it = FALSE)
points(qqScaled$x, qqScaled$y, col = "red", cex = 0.6, pch = 20);
abline(a=0, b=1, col = "blue")
legend("topleft", legend = c("Unscaled TOM", "Scaled TOM"), pch = 20, col = c("black", "red"))
9共识拓扑重叠的计算
计算完成后,假设我们有一个基因在两个数据集中表达,如何认为它是差异基因呢 ️❓
根据WGCNA中的解释,如果这个基因在两个数据集中差异都很大,这个时候我们才认为它是一个变化差异大的基因,就像达成共识一样。😜
consensusTOM = pmin(TOM[1, , ], TOM[2, , ])
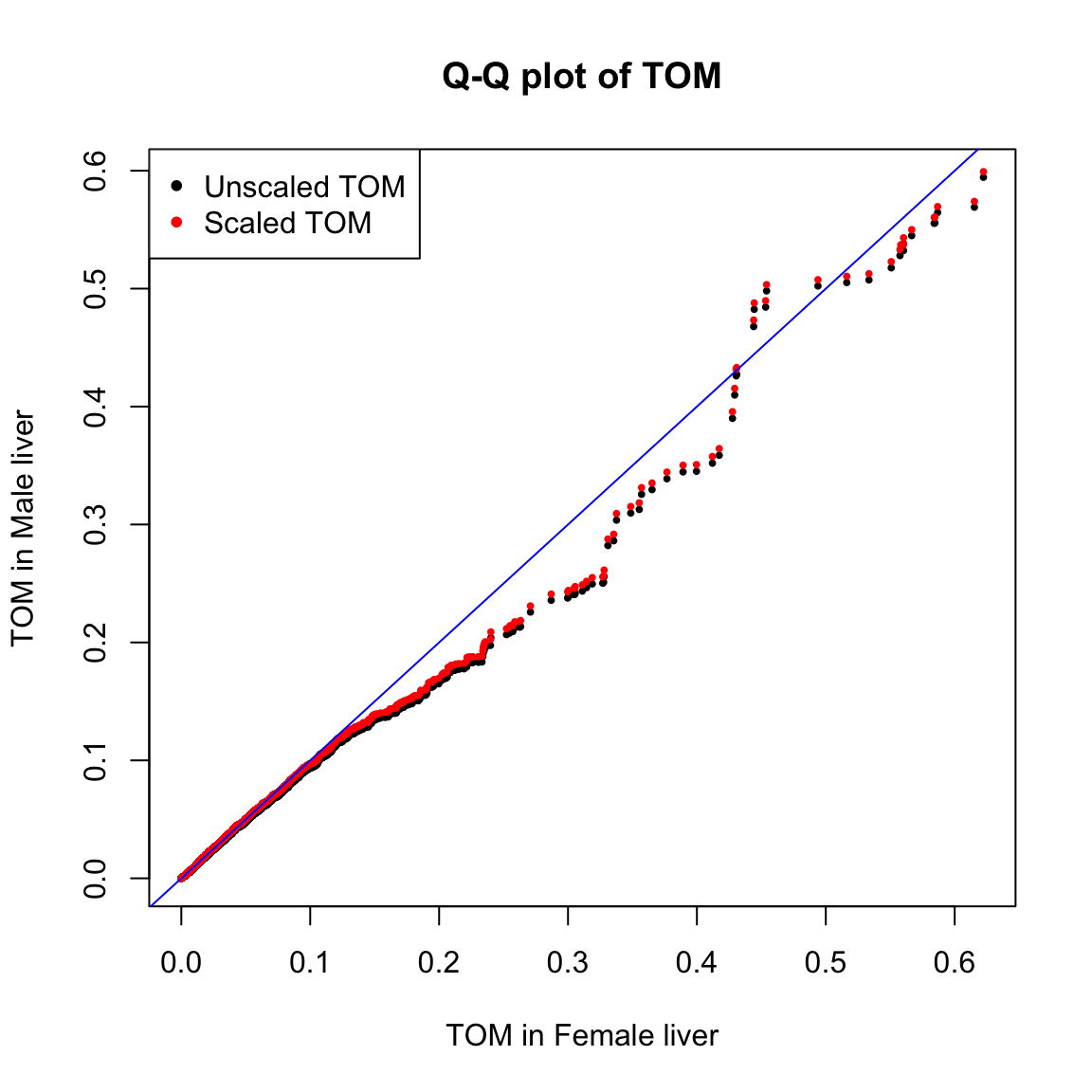
10聚类与模块识别
10.1 计算模块
consTree <- hclust(as.dist(1-consensusTOM), method = "average")
minModuleSize <- 30
unmergedLabels <- cutreeDynamic(dendro = consTree, distM = 1-consensusTOM,
deepSplit = 2, cutHeight = 0.995,
minClusterSize = minModuleSize,
pamRespectsDendro = F)
unmergedColors <- labels2colors(unmergedLabels)
table(unmergedLabels)

10.2 可视化
sizeGrWindow(8,6)
plotDendroAndColors(consTree, unmergedColors, "Dynamic Tree Cut",
dendroLabels = F, hang = 0.03,
addGuide = T, guideHang = 0.05)
10.3 计算模块MEs
可以用于后面合并相似模块哦。🤠
unmergedMEs <- multiSetMEs(multiExpr, colors = NULL, universalColors = unmergedColors)
consMEDiss <- consensusMEDissimilarity(unmergedMEs)
consMETree <- hclust(as.dist(consMEDiss), method = "average")

10.4 可视化聚类结果
sizeGrWindow(7,6)
par(mfrow = c(1,1))
plot(consMETree, main = "Consensus clustering of consensus module eigengenes",
xlab = "", sub = "")
abline(h=0.25, col = "red")
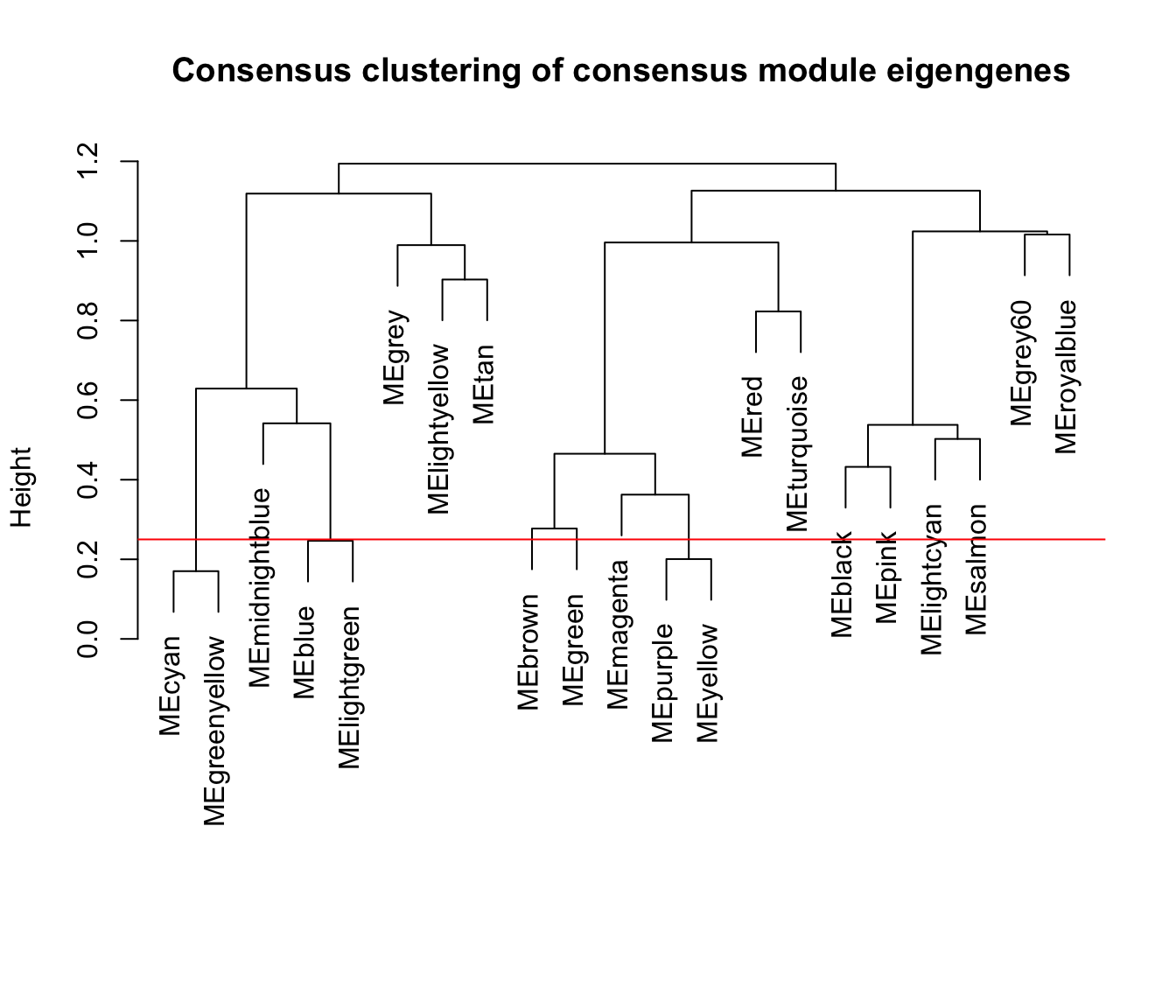
10.5 合并
merge <- mergeCloseModules(multiExpr, unmergedLabels, cutHeight = 0.25, verbose = 3)
10.6 提取重要数据
moduleLabels <- merge$colors
moduleColors <- labels2colors(moduleLabels)
consMEs <- merge$newMEs
10.7 最终可视化
sizeGrWindow(9,6)
plotDendroAndColors(consTree, cbind(unmergedColors, moduleColors),
c("Unmerged", "Merged"),
dendroLabels = F, hang = 0.03,
addGuide = T, guideHang = 0.05)
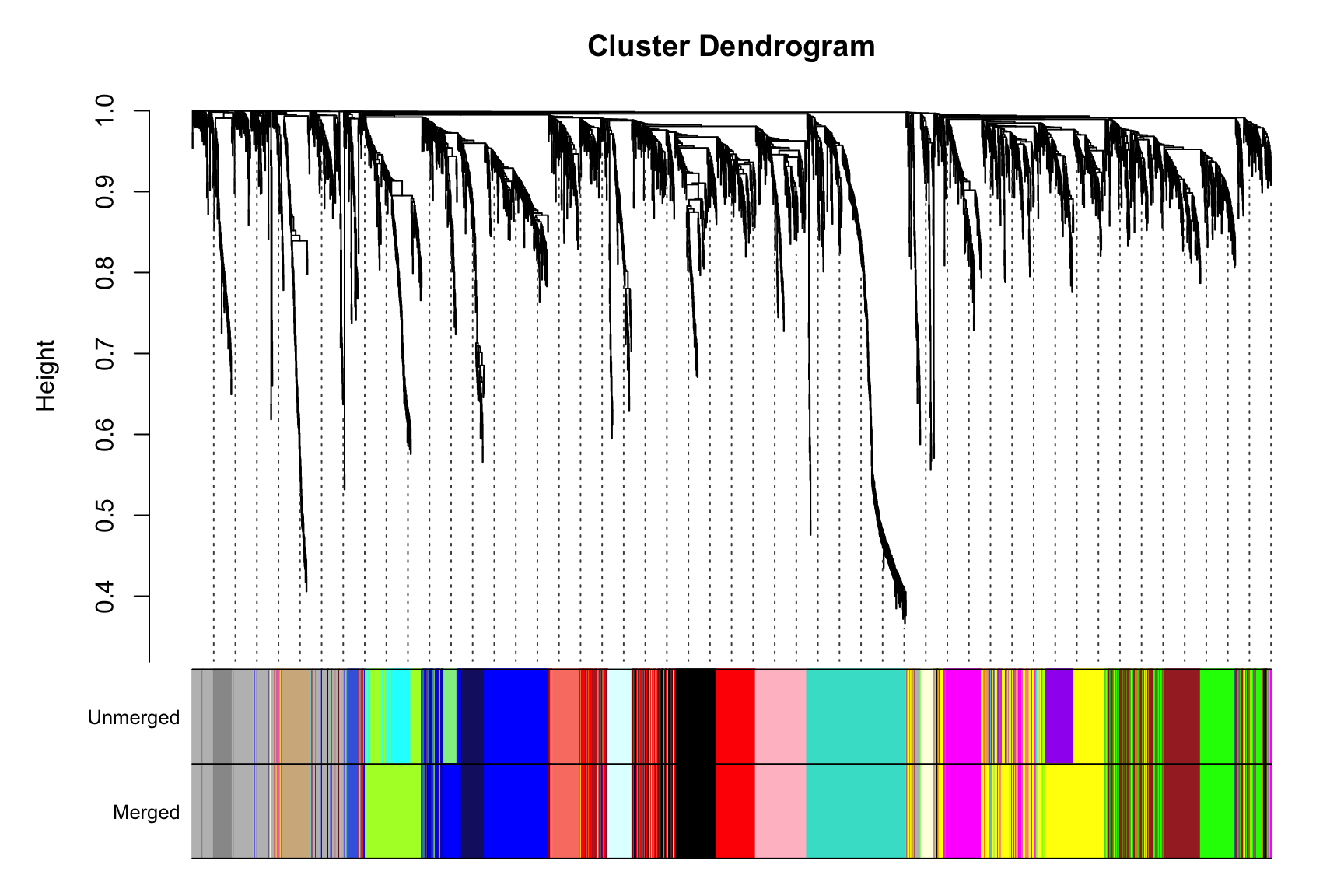
11Save一下
老规矩,保存一下我们辛辛苦苦运行的数据吧。🎩
save(consMEs, moduleColors, moduleLabels, consTree, file = "./Consensus-NetworkConstruction-man.RData")

点个在看吧各位~ ✐.ɴɪᴄᴇ ᴅᴀʏ 〰
📍 🤩 WGCNA | 值得你深入学习的生信分析方法!~
📍 🤩 ComplexHeatmap | 颜狗写的高颜值热图代码!
📍 🤥 ComplexHeatmap | 你的热图注释还挤在一起看不清吗!?
📍 🤨 Google | 谷歌翻译崩了我们怎么办!?(附完美解决方案)
📍 🤩 scRNA-seq | 吐血整理的单细胞入门教程
📍 🤣 NetworkD3 | 让我们一起画个动态的桑基图吧~
📍 🤩 RColorBrewer | 再多的配色也能轻松搞定!~
📍 🧐 rms | 批量完成你的线性回归
📍 🤩 CMplot | 完美复刻Nature上的曼哈顿图
📍 🤠 Network | 高颜值动态网络可视化工具
📍 🤗 boxjitter | 完美复刻Nature上的高颜值统计图
📍 🤫 linkET | 完美解决ggcor安装失败方案(附教程)
📍 ......
本文由 mdnice 多平台发布