一、Description

One measure of unsortedness'' in a sequence is the number of pairs of entries that are out of order with respect to each other. For instance, in the letter sequence DAABEC’‘, this measure is 5, since D is greater than four letters to its right and E is greater than one letter to its right. This measure is called the number of inversions in the sequence. The sequence AACEDGG'' has only one inversion (E and D)---it is nearly sorted---while the sequence ZWQM’’ has 6 inversions (it is as unsorted as can be—exactly the reverse of sorted).
You are responsible for cataloguing a sequence of DNA strings (sequences containing only the four letters A, C, G, and T). However, you want to catalog them, not in alphabetical order, but rather in order of sortedness'', from most sorted’’ to ``least sorted’'. All the strings are of the same length.
二、Input
The first line contains two integers: a positive integer n (0 < n <= 50) giving the length of the strings; and a positive integer m (0 < m <= 100) giving the number of strings. These are followed by m lines, each containing a string of length n.
三、Output
Output the list of input strings, arranged from most sorted'' to least sorted’'. Since two strings can be equally sorted, then output them according to the orginal order.
四、Sample Input
10 6
AACATGAAGG
TTTTGGCCAA
TTTGGCCAAA
GATCAGATTT
CCCGGGGGGA
ATCGATGCAT
五、Sample Output
CCCGGGGGGA
AACATGAAGG
GATCAGATTT
ATCGATGCAT
TTTTGGCCAA
TTTGGCCAAA
六、Source
#include <stdio.h>
#include <stdlib.h>
#include <string.h>
#define DNA_LEN 50
#define DNA_NUM 100
#define BUFFER_SIZE 10000
typedef struct
{
int unsortedness;
char dnaString[DNA_LEN];
}Dna;
typedef struct
{
int dnaLen;
int dnaNum;
Dna dna[DNA_NUM];
Dna* pDna[DNA_NUM];
}DnaSequence;
DnaSequence DnaSeq;
void GetDnaSequence(DnaSequence *dnaSeq)
{
int i;
scanf("%d %d\n", &dnaSeq->dnaLen, &dnaSeq->dnaNum);
for(i = 0; i < dnaSeq->dnaNum; i++)
{
if(NULL == gets(dnaSeq->dna[i].dnaString)) break;
dnaSeq->pDna[i] = &dnaSeq->dna[i];
}
}
void PrintDnaSequence(DnaSequence *dnaSeq)
{
int i;
for(i = 0; i < dnaSeq->dnaNum; i++)
{
printf("%s\n", dnaSeq->pDna[i]->dnaString);
}
}
/*
void CalcUnsortedness(Dna* dna, int dnaLen)
{
int delta,i,j;
dna->unsortedness = 0;
for(i = 0; i < dnaLen; i++)
{
for(j = i+1; j < dnaLen; j++)
{
delta = dna->dnaString[i] - dna->dnaString[j];
if(delta > 0) dna->unsortedness++;
}
}
}
*/
void CalcUnsortedness(Dna* dna, int dnaLen)
{
int i;
int A = 0, C = 0, G = 0;
dna->unsortedness = 0;
for(i = dnaLen - 1; i >= 0; i--)
{
switch(dna->dnaString[i])
{
case 'A':
A++;
break;
case 'C':
C++;
dna->unsortedness += A;
break;
case 'G':
G++;
dna->unsortedness += A+C;
break;
case 'T':
dna->unsortedness += A+C+G;
break;
default:
break;
}
}
}
int SortCmp(const void* elem1, const void* elem2)
{
Dna* dna1 = (Dna *)(*(size_t*)elem1);
Dna* dna2 = (Dna *)(*(size_t*)elem2);
return dna1->unsortedness - dna2->unsortedness;
}
char g_mergeBuffer[BUFFER_SIZE];
void Merge(char* array, int elemSize, int left, int mid, int right, int (*SortCmp)(const void*, const void*))
{
int i = left;
int j = mid;
int bufIdx = 0;
while(i < mid && j <= right)
{
if(SortCmp(&array[i*elemSize], &array[j*elemSize]) <= 0)
{
memcpy(&g_mergeBuffer[bufIdx], &array[i*elemSize], elemSize);
i++;
}
else
{
memcpy(&g_mergeBuffer[bufIdx], &array[j*elemSize], elemSize);
j++;
}
bufIdx += elemSize;
}
for(; i < mid; i++)
{
memcpy(&g_mergeBuffer[bufIdx], &array[i*elemSize], elemSize);
bufIdx += elemSize;
}
for(; j <= right; j++)
{
memcpy(&g_mergeBuffer[bufIdx], &array[j*elemSize], elemSize);
bufIdx += elemSize;
}
memcpy(&array[left*elemSize], g_mergeBuffer, (right-left+1)*elemSize);
}
void MergeSort(void* array, int arrayLen, int elemSize, int (*SortCmp)(const void*, const void*))
{
int loop, left, mid, right = 0;
for(loop = 1; loop < arrayLen; loop *= 2)
{
left = 0;
right = 0;
while(right < arrayLen - 1)
{
mid = left + loop;
right = (mid + loop - 1 > arrayLen - 1) ? (arrayLen - 1) : (mid + loop - 1);
Merge((char*)array, elemSize, left, mid, right, SortCmp);
left = left + loop * 2;
}
}
}
void ProcDnaSequence(DnaSequence *dnaSeq)
{
int i;
int elemSize = sizeof(dnaSeq->pDna[0]);
for(i = 0; i < dnaSeq->dnaNum; i++)
{
CalcUnsortedness(&dnaSeq->dna[i], dnaSeq->dnaLen);
}
MergeSort(dnaSeq->pDna, dnaSeq->dnaNum, elemSize, SortCmp);
}
int main()
{
GetDnaSequence(&DnaSeq);
ProcDnaSequence(&DnaSeq);
PrintDnaSequence(&DnaSeq);
return 0;
}
七、总结
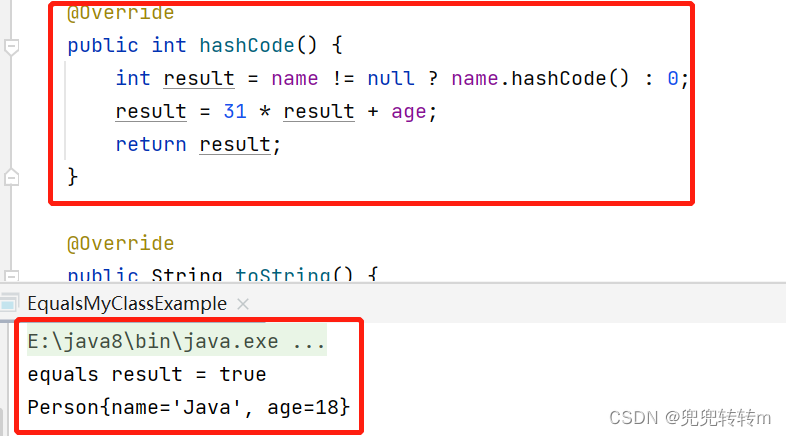
逆序数可以用来描述一个序列混乱程度的量。例如,“DAABEC”的逆序数为5,其中D大于他右边的4个数,E大于他右边的1个数,4+1=5;又如,“ZWQM”的逆序数为3+2+1+0=6。现在有许多长度一样的字符串,每个字符串里面只会出现四种字母(A,T,G,C)。要求编写程序,将这些字符串按照他们的逆序数进行排序。
要求用稳定的排序算法,所以选择了归并排序。计算逆序数原本没想太多用的暴力遍历,但是后来看评论,发现一种有趣的算法。

![[附源码]Python计算机毕业设计儿童闲置物品交易网站](https://img-blog.csdnimg.cn/8085f7fcbf1c49688ca327ba9ce8c968.png)