这个例子展示了如何使用coregistration函数通过脚本自动完成MEG-MRI的coregistration。一般情况下,该方法的结果与人工共配准的结果是一致的。
**但一定要注意:**协同配准的质量在很大程度上取决于受试者准备过程中收集的头形点(HSP)的质量和t1加权MRI的质量。请谨慎使用并检查coregistration错误。
# Author: Jon Houck <jon.houck@gmail.com>
# Guillaume Favelier <guillaume.favelier@gmail.com>
#
# License: BSD-3-Clause
# Copyright the MNE-Python contributors.
import numpy as np
import mne
from mne.coreg import Coregistration
from mne.io import read_info
data_path = mne.datasets.sample.data_path()
# data_path and all paths built from it are pathlib.Path objects
subjects_dir = data_path / "subjects"
subject = "sample"
fname_raw = data_path / "MEG" / subject / f"{subject}_audvis_raw.fif"
info = read_info(fname_raw)
plot_kwargs = dict(
subject=subject,
subjects_dir=subjects_dir,
surfaces="head-dense",
dig=True,
eeg=[],
meg="sensors",
show_axes=True,
coord_frame="meg",
)
view_kwargs = dict(azimuth=45, elevation=90, distance=0.6, focalpoint=(0.0, 0.0, 0.0))
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
设置配准模型
fiducials = "estimated" # get fiducials from fsaverage
coreg = Coregistration(info, subject, subjects_dir, fiducials=fiducials)
fig = mne.viz.plot_alignment(info, trans=coreg.trans, **plot_kwargs)

Triangle neighbors and vertex normals... Using high resolution head model in /home/circleci/mne_data/MNE-sample-data/subjects/sample/surf/lh.seghead
Triangle neighbors and vertex normals...
Estimating fiducials from fsaverage.
Using lh.seghead for head surface.
Channel types:: grad: 203, mag: 102
粗基准匹配
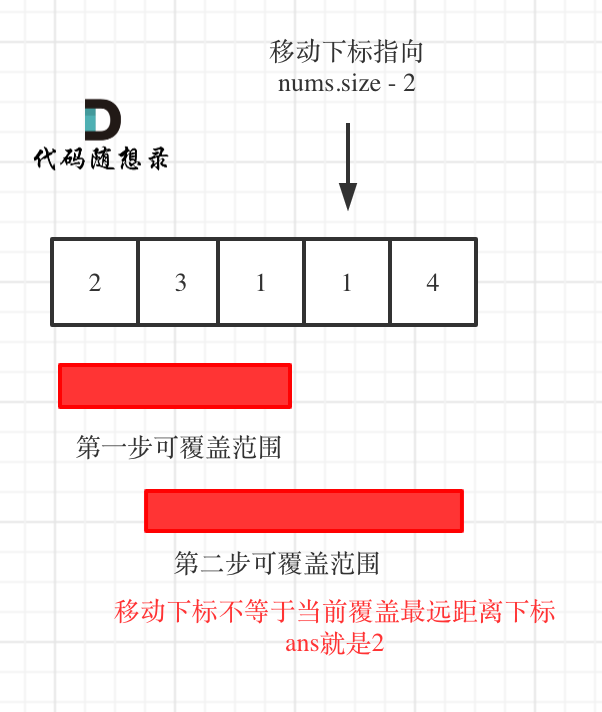
首先使用3个基准点进行协同配准拟合。这允许在使用头形点进一步优化之前找到一个良好的初始解决方案。这也可以用于检测离皮肤表面太远的异常头部形状点。例如,可以看到在这个数据集上有一个这样的点,我们将在后续的拟合中忽略它。
coreg.fit_fiducials(verbose=True)
fig = mne.viz.plot_alignment(info, trans=coreg.trans, **plot_kwargs)

Aligning using fiducials
Start median distance: 17.29 mm
End median distance: 11.11 mm
Using lh.seghead for head surface.
Channel types:: grad: 203, mag: 102
用ICP进行改进
接下来,我们使用迭代最近点(ICP)算法的多次迭代来完善转换。由于初始基准是从fsaaverage中获得,而不是从GUI中的精确手动选择中获得,因此我们为nasion做了一个减轻权重的拟合。
coreg.fit_icp(n_iterations=6, nasion_weight=2.0, verbose=True)
fig = mne.viz.plot_alignment(info, trans=coreg.trans, **plot_kwargs)

Aligning using ICP
Start median distance: 11.11 mm
ICP 1 median distance: 6.57 mm
ICP 2 median distance: 4.39 mm
ICP 3 median distance: 3.48 mm
ICP 4 median distance: 2.86 mm
ICP 5 median distance: 2.43 mm
ICP 6 median distance: 2.14 mm
End median distance: 2.14 mm
Using lh.seghead for head surface.
Channel types:: grad: 203, mag: 102
排除坏的点
现在很明显,我们有一个点是异常值,应该被删除。
coreg.omit_head_shape_points(distance=5.0 / 1000) # distance is in meters
Coregistration: Excluding 3 head shape points with distance >= 0.005 m.
coreg.fit_icp(n_iterations=20, nasion_weight=10.0, verbose=True)
fig = mne.viz.plot_alignment(info, trans=coreg.trans, **plot_kwargs)
mne.viz.set_3d_view(fig, **view_kwargs)
dists = coreg.compute_dig_mri_distances() * 1e3 # in mm
print(
f"Distance between HSP and MRI (mean/min/max):\n{np.mean(dists):.2f} mm "
f"/ {np.min(dists):.2f} mm / {np.max(dists):.2f} mm"
)
最终配准
coreg.fit_icp(n_iterations=20, nasion_weight=10.0, verbose=True)
fig = mne.viz.plot_alignment(info, trans=coreg.trans, **plot_kwargs)
mne.viz.set_3d_view(fig, **view_kwargs)
dists = coreg.compute_dig_mri_distances() * 1e3 # in mm
print(
f"Distance between HSP and MRI (mean/min/max):\n{np.mean(dists):.2f} mm "
f"/ {np.min(dists):.2f} mm / {np.max(dists):.2f} mm"
)

Aligning using ICP
Start median distance: 2.00 mm
ICP 1 median distance: 1.99 mm
ICP 2 median distance: 2.02 mm
ICP 3 median distance: 1.79 mm
ICP 4 median distance: 1.78 mm
ICP 5 median distance: 1.77 mm
End median distance: 1.77 mm
Using lh.seghead for head surface.
Channel types:: grad: 203, mag: 102
Distance between HSP and MRI (mean/min/max):
2.00 mm / 0.19 mm / 6.37 mm
别忘了保存得到的trans矩阵!
mne.write_trans('/path/to/filename-trans.fif', coreg.trans)
mne.coreg.coregistration能够使用set_scale_mode()计算MRI比例因子,这对于创建替代的MRI受试者很有用,即使用与受试者头部数字化匹配的模板MRI,例如mne.datasets.fetch_infant_template()。当需要缩放时,应该使用mne.scale_mri()创建缩放的替代的MRI。