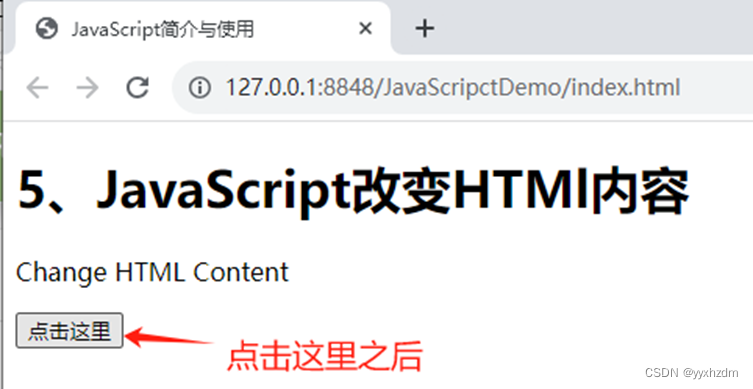
大家好,今天我们分享的是单细胞的学习教程https://www.singlecellworkshop.com/analysis-tutorial.html 教程的作者使用了四个样本,但是没有使用harmony或者其他方法去整合 去除批次效应。
主要内容:
-
SCTransform流程代码及结果
-
harmony流程代码及结果
-
seurat单样本标准流程代码及结果
-
三种方法结果比较
是不是这四个样本就不需要去批次效应呢?接下来我们探索一下
1 首先是把教程的代码跑一遍
# load Seurat package
library(Seurat)
dir.create("~/gzh/harmony_sct",recursive = TRUE)
setwd("~/gzh/harmony_sct/")
getwd()
list.files()
# 1 不去除批次效应,教程的步骤----
{
pfc2.data <- Read10X(data.dir = "raw-data/pfc-sample2")
pfc3.data <- Read10X(data.dir = "raw-data/pfc-sample3")
pfc5.data <- Read10X(data.dir = "raw-data/pfc-sample5")
pfc7.data <- Read10X(data.dir = "raw-data/pfc-sample7")
# create a new Seurat object for each sample
# min.cells = 3, only genes detected in at least 3 cells will be included
# min.features = 200, only cells with at least 200 genes detected will be included
pfc2 <- CreateSeuratObject(counts = pfc2.data, project = "pfc-demo", min.cells = 3, min.features = 200)
pfc3 <- CreateSeuratObject(counts = pfc3.data, project = "pfc-demo", min.cells = 3, min.features = 200)
pfc5 <- CreateSeuratObject(counts = pfc5.data, project = "pfc-demo", min.cells = 3, min.features = 200)
pfc7 <- CreateSeuratObject(counts = pfc7.data, project = "pfc-demo", min.cells = 3, min.features = 200)
pfc2
# remove the raw data to save processing space
rm(pfc2.data, pfc3.data, pfc5.data, pfc7.data)
# inspect the metadata for one of our objects using the 'head' function
head(pfc2@meta.data)
pfc2@meta.data$sample_number <- "2"
pfc3@meta.data$sample_number <- "3"
pfc5@meta.data$sample_number <- "5"
pfc7@meta.data$sample_number <- "7"
# merge multiple Seurat objects
pfc <- merge(x = pfc2, y = list(pfc3, pfc5, pfc7))
# remove individual objects to save processing space
rm(pfc2, pfc3, pfc5, pfc7)
# inpect our new combined object
pfc
# an important metadata slot to add in every experiment is the ratio of mitochondrial genes
# detected in each cell - this can be used as a proxy for cell quality in most preparations
pfc[["percent.mt"]] <- PercentageFeatureSet(object = pfc, pattern = "^mt-")
# percent.mt cutoffs typically range from 5-10% depending on the sample
VlnPlot(pfc, features = c("percent.mt"), pt.size=0, y.max=15)
pfc <- subset(pfc, subset = percent.mt < 5)
VlnPlot(pfc, features = c("nFeature_RNA"), pt.size=0, y.max=2000)
pfc <- subset(pfc, subset = nFeature_RNA > 600)
# inspect our QC metrics again
VlnPlot(pfc, features = c("nFeature_RNA","nCount_RNA","percent.mt"), pt.size=0)
# this may take several minutes to execute, and progress will display in the console
pfc <- SCTransform(pfc)
pfc <- RunPCA(pfc, npcs = 60)
pfc <- FindNeighbors(pfc, dims = 1:60)
pfc <- FindClusters(pfc, resolution = 0.7)
pfc <- RunUMAP(pfc, dims = 1:60)
DimPlot(pfc, label=T)
DimPlot(pfc, group.by="sample_number")
}
DefaultAssay(pfc)
Assays(pfc)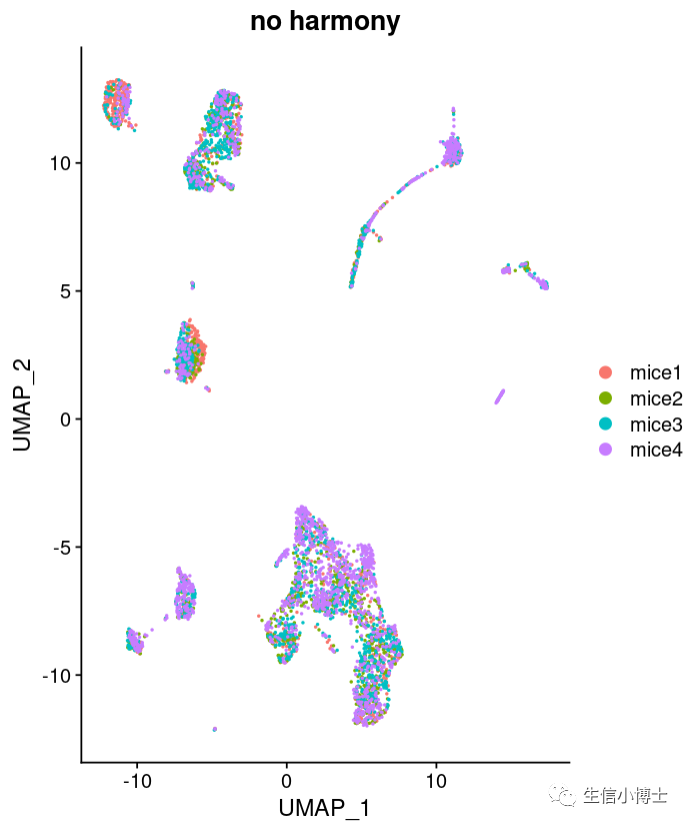
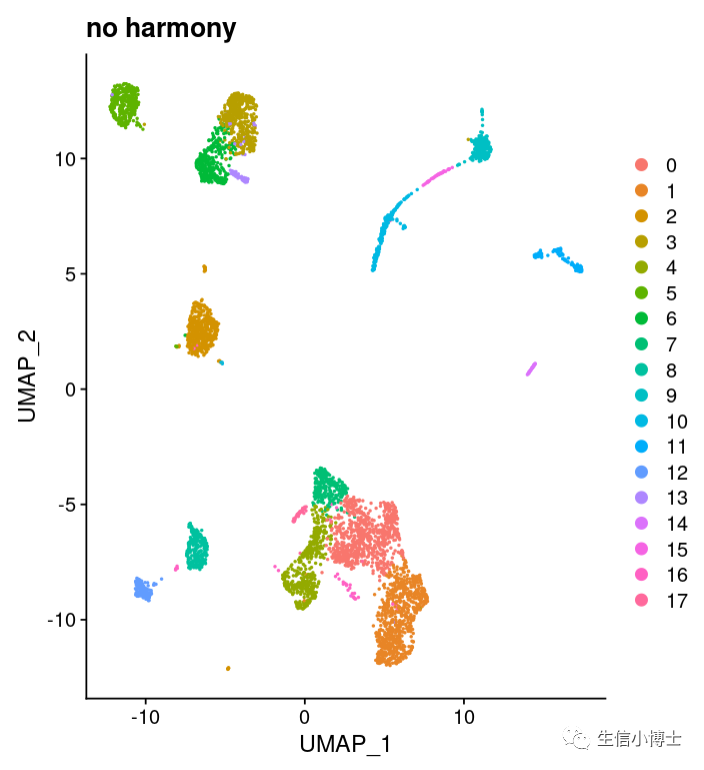
我们可以看到就算不去批次效应,结果也挺好的
02
2 然后使用harmony去除批次效应 看看效果
subset_data=pfc
DefaultAssay(subset_data)='RNA'
{
library(dplyr)
DimPlot(subset_data)
subset_data[["percent.mt"]] <- PercentageFeatureSet(subset_data, pattern = "^mt-")
VlnPlot(subset_data, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
subset_data = subset_data %>%
Seurat::NormalizeData(verbose = FALSE) %>%
FindVariableFeatures(selection.method = "vst", nfeatures = 2000) %>%
ScaleData(verbose = FALSE) %>%
RunPCA(npcs = 30, verbose = FALSE)
ElbowPlot(subset_data, ndims = 30)
VlnPlot(subset_data, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
head(subset_data@meta.data)
library(stringr)
table(str_split(colnames(subset_data),pattern = "_",simplify = TRUE)[,2])
subset_data@meta.data$stim <-paste0('mice',str_split(colnames(subset_data),pattern = "_",simplify = TRUE)[,2])
#table(subset_data$stim)
library('harmony')
subset_data <- subset_data %>% RunHarmony("stim", plot_convergence = TRUE)
harmony_embeddings <- Embeddings(subset_data, 'harmony')
#######################cluster
dims = 1:30
subset_data <- subset_data %>%
RunUMAP(reduction = "harmony", dims = dims) %>%
RunTSNE(reduction = "harmony", dims = dims) %>%
FindNeighbors(reduction = "harmony", dims = dims)
subset_data=FindClusters(subset_data,resolution =0.7)
DimPlot(subset_data,group)
head(subset_data@meta.data)
head(pfc@meta.data)
}同样的分辨率下对比两次的结果
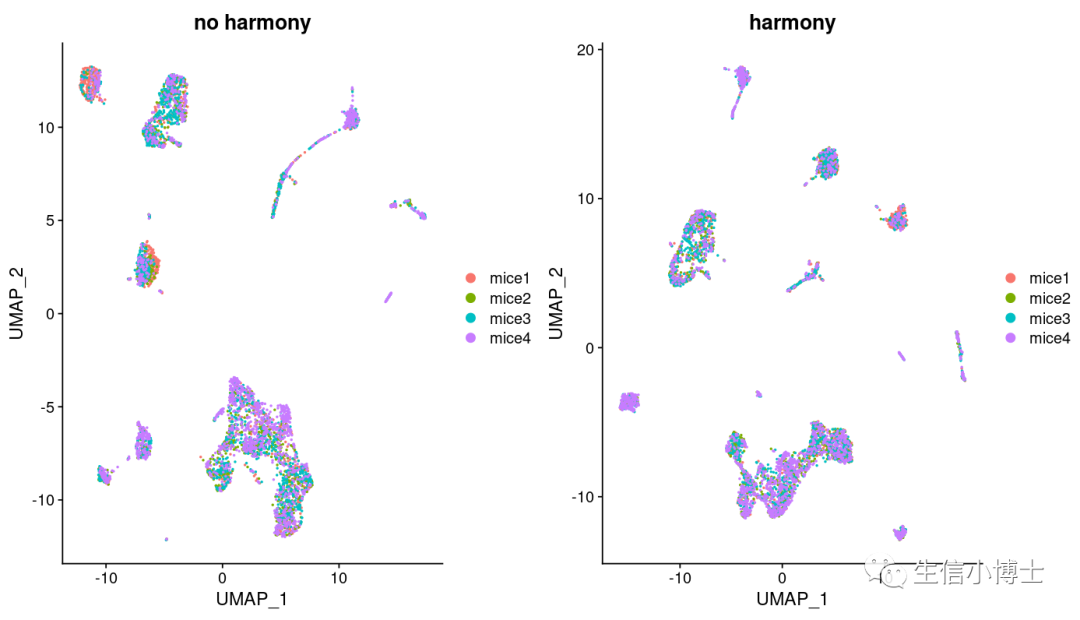
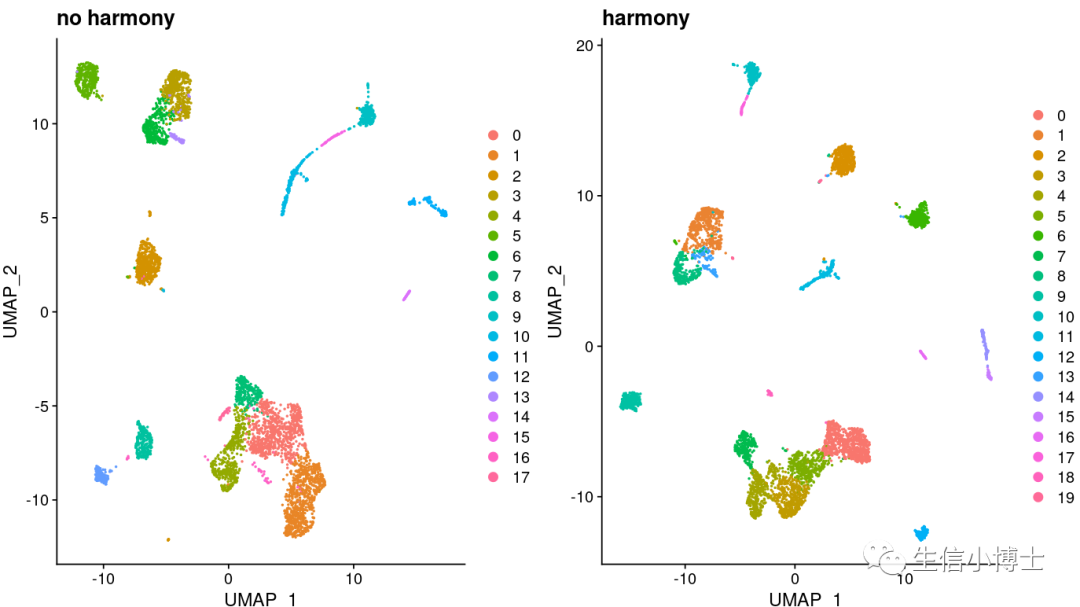
我们发现hamony之后多了两个亚群,但是样本总体分布好像影响并不大。所以我们看下harmony前后,每个亚群中的细胞数量变化
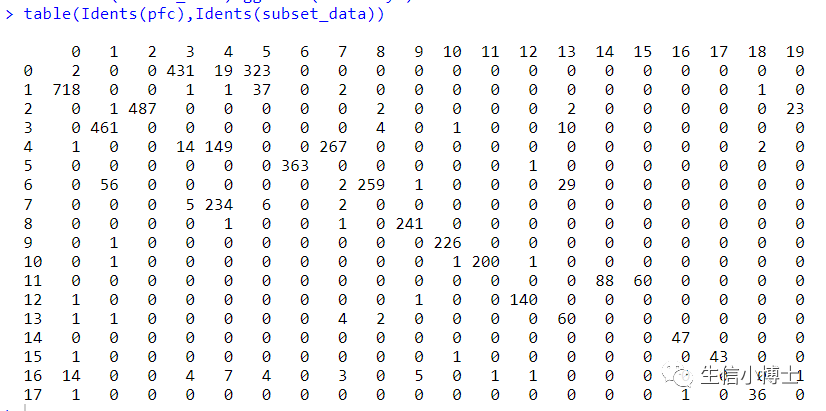
总体看上去,harmony前后对大多数亚群影响并不大,且harmony前后有很多亚群多是可以互相重合的。(个人觉得之所以教程作者不去除批次效应是因为他所选的四个样本都是control组)

如果想更详细的比较harmony前后的变化,我们可以从细胞命名、差异分析、拟时序分析等等结果来看啦~
那为什么作者只是使用了SCTtransform这个函数就可以?达到这么好的效果,是不是SCTtransform可以去除批次效应?——当然不是,不信你去看官网~

3 .不死心的话,我们不使用SCTtransform,也不去除批次效应,只使用seurat标准流程试试
3#3 标准流程-----
head(subset_data@meta.data)
All=CreateSeuratObject(counts = subset_data@assays$RNA@counts,meta.data = subset_data@meta.data)
{
VlnPlot(All, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
All = All%>%Seurat::NormalizeData(verbose = FALSE) %>%
FindVariableFeatures(selection.method = "vst" ) %>%
ScaleData(verbose = FALSE)
All = RunPCA(All, npcs = 30, verbose = FALSE)
ElbowPlot(All, ndims = 30)
#######################cluster
All <- All %>%
RunUMAP(reduction = "pca", dims = 1:30) %>%
RunTSNE(reduction = "pca", dims = 1:30) %>%
FindNeighbors(reduction = "pca", dims = 1:30)
All<-All%>% FindClusters(resolution = 0.7) %>% identity()
DimPlot(All,group.by ='stim' )+ggtitle("standard_pipeline")
}
head(All@meta.data)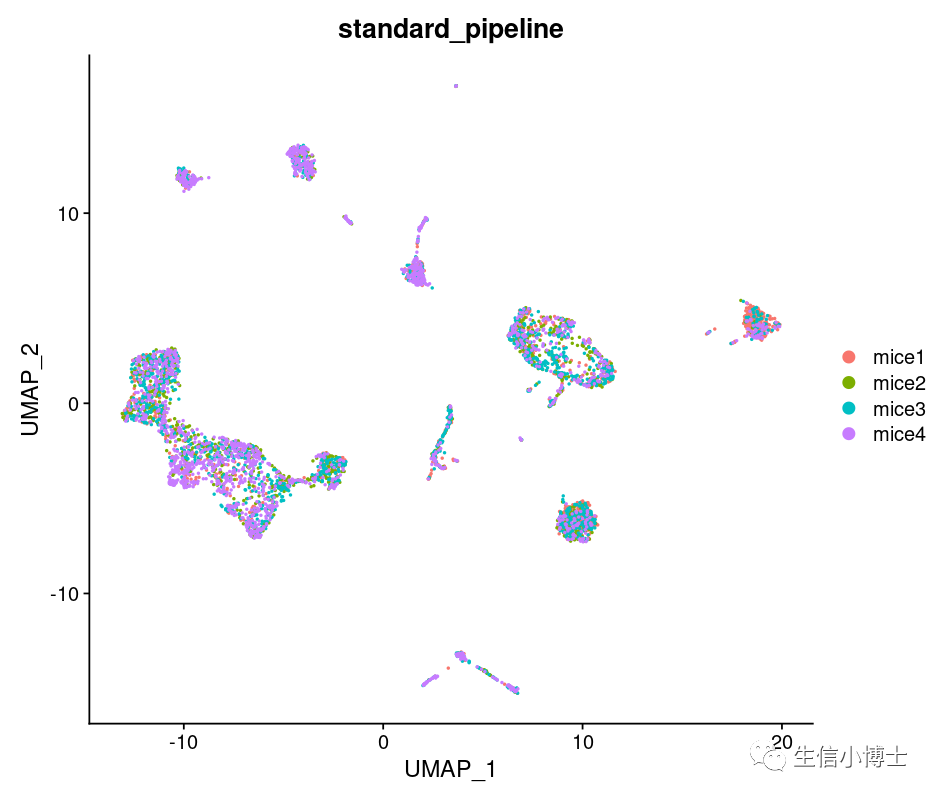
结果看上去也还可以吧
我们对比一下三种方法:
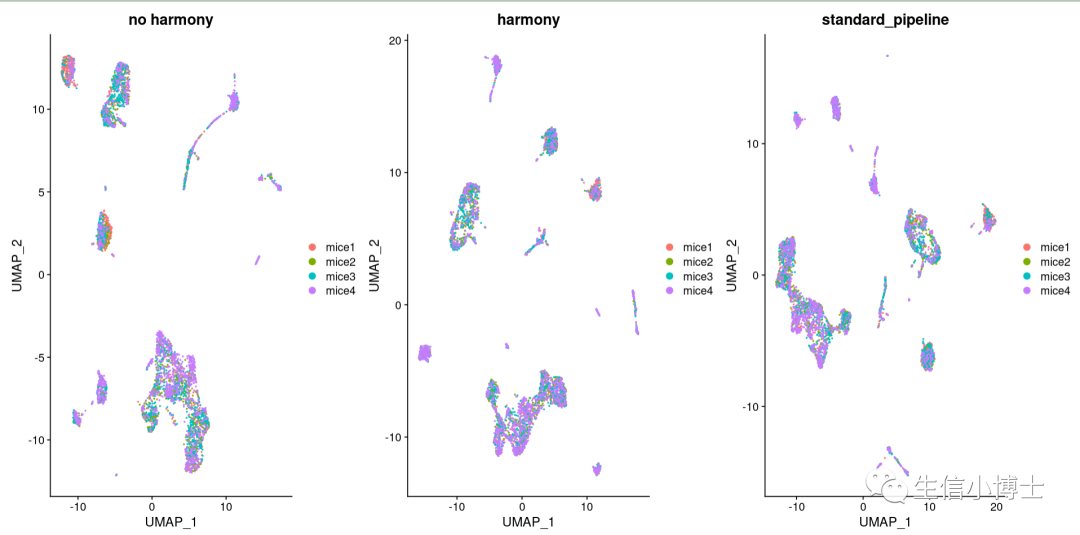
肉眼看不出来有多大区别吧
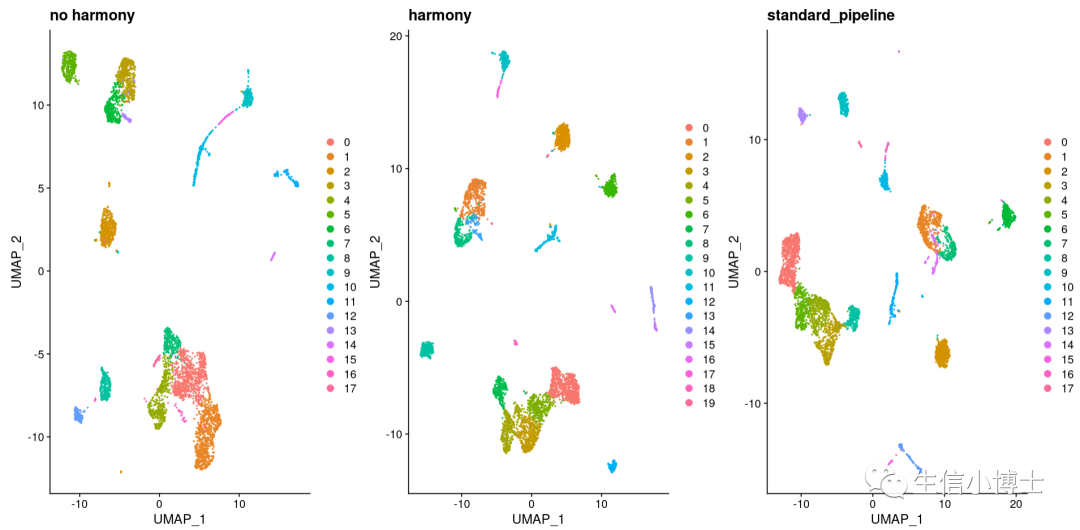
有意思的是sctransform和标准流程都能得到17个亚群
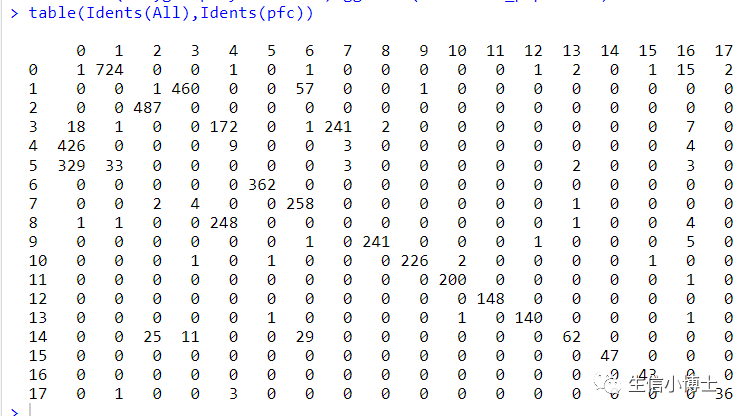
所以大家觉得我们需要harmnoy去除批次效应吗?
4 结论: 虽然不去批次效应也能拿到很好的结果,个人还是建议使用harmony,你觉得呢?
尽管在某些情况下即使不去除批次效应也能得到看似合理的结果,但这可能是偶然的,并且存在风险。批次效应可能掩盖或模拟出实际的生物信号,导致错误的生物学结论。因此,建议使用诸如Harmony这样的算法来校正批次效应,以提高数据分析的可靠性和准确性。
Harmony是一种用于多组数据整合的算法,它可以在保留生物学变异的同时,减少技术变异,特别是批次效应。通过对数据进行校正,Harmony可以帮助研究者更好地识别真实的生物学差异,而不是由实验条件引起的差异。
如果想更详细的比较harmony前后的变化,我们可以从细胞命名、差异分析、拟时序分析等等结果来看啦~如果大家对这个话题感兴趣我们可以开个直播聊聊~
微信公众号:生信小博士
感谢Jimmy老师的分享,此次推文的灵感来自Jimmy老师
猜你可能感兴趣:seurat个性化细胞注释并把细分亚群放回总群

![]()
看完记得顺手点个“在看”哦!