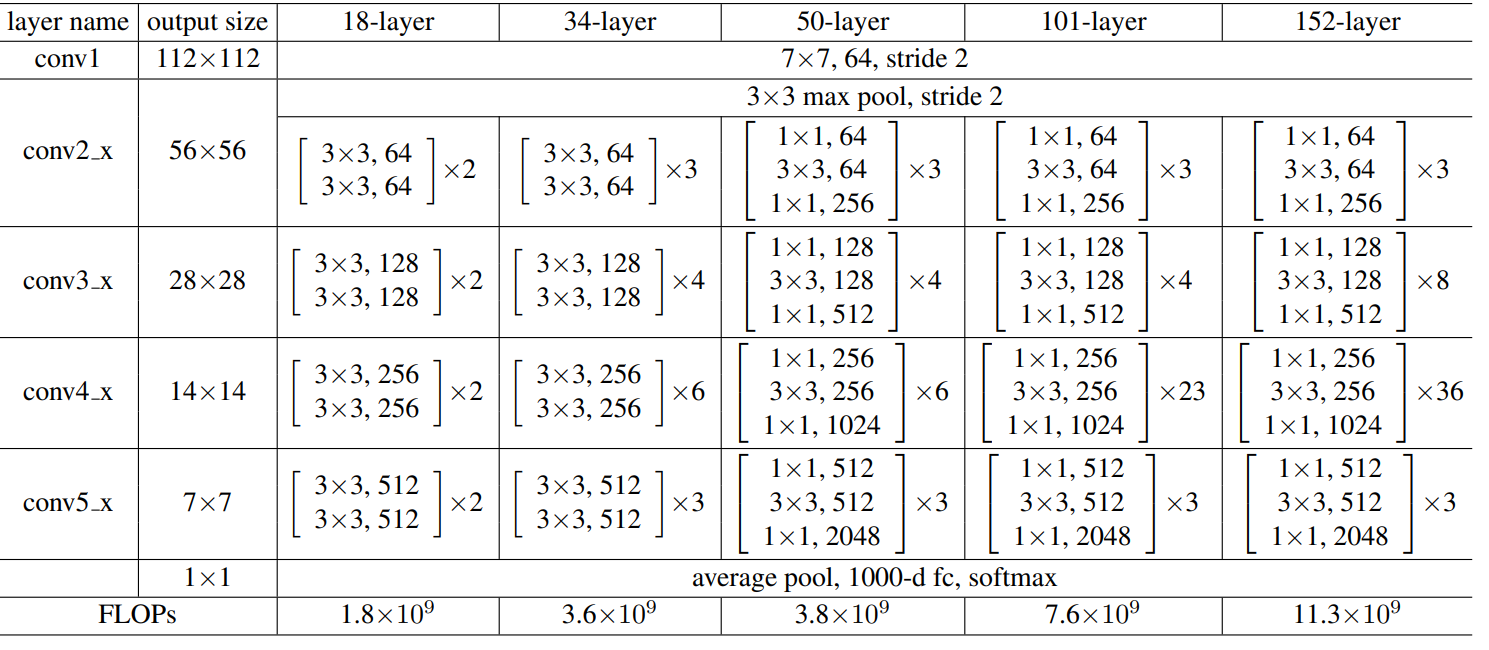
NGCF
论文地址
NGCF模型全部代码
import torch
import torch.nn as nn
import torch.nn.functional as F
class NGCF(nn.Module):
def __init__(self, n_user, n_item, norm_adj, args):
super(NGCF, self).__init__()
self.n_user = n_user
self.n_item = n_item
self.device = args.device
self.emb_size = args.embed_size
self.batch_size = args.batch_size
self.node_dropout = args.node_dropout[0]
self.mess_dropout = args.mess_dropout
self.batch_size = args.batch_size
self.norm_adj = norm_adj
self.layers = eval(args.layer_size)
self.decay = eval(args.regs)[0]
"""
*********************************************************
Init the weight of user-item.
"""
self.embedding_dict, self.weight_dict = self.init_weight()
"""
*********************************************************
Get sparse adj.
"""
self.sparse_norm_adj = self._convert_sp_mat_to_sp_tensor(self.norm_adj).to(self.device)
def init_weight(self):
# xavier init
initializer = nn.init.xavier_uniform_
embedding_dict = nn.ParameterDict({
'user_emb': nn.Parameter(initializer(torch.empty(self.n_user,
self.emb_size))),
'item_emb': nn.Parameter(initializer(torch.empty(self.n_item,
self.emb_size)))
})
weight_dict = nn.ParameterDict()
layers = [self.emb_size] + self.layers
for k in range(len(self.layers)):
weight_dict.update({'W_gc_%d'%k: nn.Parameter(initializer(torch.empty(layers[k],
layers[k+1])))})
weight_dict.update({'b_gc_%d'%k: nn.Parameter(initializer(torch.empty(1, layers[k+1])))})
weight_dict.update({'W_bi_%d'%k: nn.Parameter(initializer(torch.empty(layers[k],
layers[k+1])))})
weight_dict.update({'b_bi_%d'%k: nn.Parameter(initializer(torch.empty(1, layers[k+1])))})
return embedding_dict, weight_dict
def _convert_sp_mat_to_sp_tensor(self, X):
coo = X.tocoo()
i = torch.LongTensor([coo.row, coo.col])
v = torch.from_numpy(coo.data).float()
return torch.sparse.FloatTensor(i, v, coo.shape)
def sparse_dropout(self, x, rate, noise_shape):
random_tensor = 1 - rate
random_tensor += torch.rand(noise_shape).to(x.device)
dropout_mask = torch.floor(random_tensor).type(torch.bool)
i = x._indices()
v = x._values()
i = i[:, dropout_mask]
v = v[dropout_mask]
out = torch.sparse.FloatTensor(i, v, x.shape).to(x.device)
return out * (1. / (1 - rate))
def create_bpr_loss(self, users, pos_items, neg_items):
pos_scores = torch.sum(torch.mul(users, pos_items), axis=1)
neg_scores = torch.sum(torch.mul(users, neg_items), axis=1)
maxi = nn.LogSigmoid()(pos_scores - neg_scores)
mf_loss = -1 * torch.mean(maxi)
# cul regularizer
regularizer = (torch.norm(users) ** 2
+ torch.norm(pos_items) ** 2
+ torch.norm(neg_items) ** 2) / 2
emb_loss = self.decay * regularizer / self.batch_size
return mf_loss + emb_loss, mf_loss, emb_loss
def rating(self, u_g_embeddings, pos_i_g_embeddings):
return torch.matmul(u_g_embeddings, pos_i_g_embeddings.t())
def forward(self, users, pos_items, neg_items, drop_flag=True):
A_hat = self.sparse_dropout(self.sparse_norm_adj,
self.node_dropout,
self.sparse_norm_adj._nnz()) if drop_flag else self.sparse_norm_adj
ego_embeddings = torch.cat([self.embedding_dict['user_emb'],
self.embedding_dict['item_emb']], 0)
all_embeddings = [ego_embeddings]
for k in range(len(self.layers)):
side_embeddings = torch.sparse.mm(A_hat, ego_embeddings)
# transformed sum messages of neighbors.
sum_embeddings = torch.matmul(side_embeddings, self.weight_dict['W_gc_%d' % k]) \
+ self.weight_dict['b_gc_%d' % k]
# bi messages of neighbors.
# element-wise product
bi_embeddings = torch.mul(ego_embeddings, side_embeddings)
# transformed bi messages of neighbors.
bi_embeddings = torch.matmul(bi_embeddings, self.weight_dict['W_bi_%d' % k]) \
+ self.weight_dict['b_bi_%d' % k]
# non-linear activation.
ego_embeddings = nn.LeakyReLU(negative_slope=0.2)(sum_embeddings + bi_embeddings)
# message dropout.
ego_embeddings = nn.Dropout(self.mess_dropout[k])(ego_embeddings)
# normalize the distribution of embeddings.
norm_embeddings = F.normalize(ego_embeddings, p=2, dim=1)
all_embeddings += [norm_embeddings]
all_embeddings = torch.cat(all_embeddings, 1)
u_g_embeddings = all_embeddings[:self.n_user, :]
i_g_embeddings = all_embeddings[self.n_user:, :]
"""
*********************************************************
look up.
"""
u_g_embeddings = u_g_embeddings[users, :]
pos_i_g_embeddings = i_g_embeddings[pos_items, :]
neg_i_g_embeddings = i_g_embeddings[neg_items, :]
return u_g_embeddings, pos_i_g_embeddings, neg_i_g_embeddings
公式1
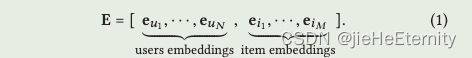
self.embedding_dict = nn.ParameterDict({
'user_emb': nn.Parameter(initializer(torch.empty(self.n_user, self.emb_size))),
'item_emb': nn.Parameter(initializer(torch.empty(self.n_item, self.emb_size)))
})
一阶传播:消息构造(公式2和3)
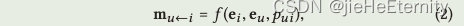
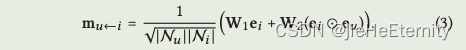
for k in range(len(self.layers)):
side_embeddings = torch.sparse.mm(A_hat, ego_embeddings)
sum_embeddings = torch.matmul(side_embeddings, self.weight_dict['W_gc_%d' % k]) + self.weight_dict['b_gc_%d' % k]
bi_embeddings = torch.mul(ego_embeddings, side_embeddings)
bi_embeddings = torch.matmul(bi_embeddings, self.weight_dict['W_bi_%d' % k]) + self.weight_dict['b_bi_%d' % k]
消息聚合 (公式 4):
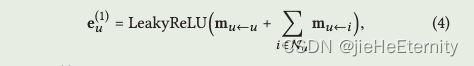
ego_embeddings = nn.LeakyReLU(negative_slope=0.2)(sum_embeddings + bi_embeddings)
高阶传播 (公式 5 和 6):
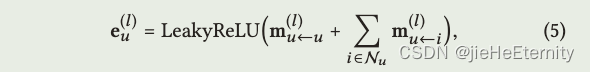
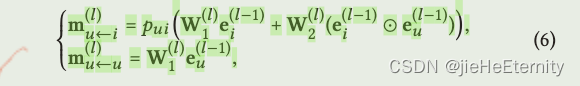
ego_embeddings = nn.Dropout(self.mess_dropout[k])(ego_embeddings)
norm_embeddings = F.normalize(ego_embeddings, p=2, dim=1)
all_embeddings += [norm_embeddings]
模型预测 (公式 9 和 10):
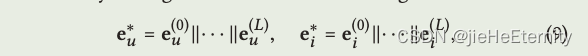
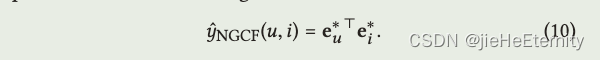
def rating(self, u_g_embeddings, pos_i_g_embeddings):
return torch.matmul(u_g_embeddings, pos_i_g_embeddings.t())
优化(公式11)
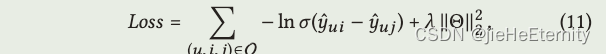
def create_bpr_loss(self, users, pos_items, neg_items):
pos_scores = torch.sum(torch.mul(users, pos_items), axis=1)
neg_scores = torch.sum(torch.mul(users, neg_items), axis=1)
maxi = nn.LogSigmoid()(pos_scores - neg_scores)
mf_loss = -1 * torch.mean(maxi)
# ... remaining code for regularizer
(2)Ligitgcn
import world
import torch
from dataloader import BasicDataset
from torch import nn
import numpy as np
class BasicModel(nn.Module):
def __init__(self):
super(BasicModel, self).__init__()
def getUsersRating(self, users):
raise NotImplementedError
class PairWiseModel(BasicModel):
def __init__(self):
super(PairWiseModel, self).__init__()
def bpr_loss(self, users, pos, neg):
"""
Parameters:
users: users list
pos: positive items for corresponding users
neg: negative items for corresponding users
Return:
(log-loss, l2-loss)
"""
raise NotImplementedError
class PureMF(BasicModel):
def __init__(self,
config:dict,
dataset:BasicDataset):
super(PureMF, self).__init__()
self.num_users = dataset.n_users
self.num_items = dataset.m_items
self.latent_dim = config['latent_dim_rec']
self.f = nn.Sigmoid()
self.__init_weight()
def __init_weight(self):
self.embedding_user = torch.nn.Embedding(
num_embeddings=self.num_users, embedding_dim=self.latent_dim)
self.embedding_item = torch.nn.Embedding(
num_embeddings=self.num_items, embedding_dim=self.latent_dim)
print("using Normal distribution N(0,1) initialization for PureMF")
def getUsersRating(self, users):
users = users.long()
users_emb = self.embedding_user(users)
items_emb = self.embedding_item.weight
scores = torch.matmul(users_emb, items_emb.t())
return self.f(scores)
def bpr_loss(self, users, pos, neg):
users_emb = self.embedding_user(users.long())
pos_emb = self.embedding_item(pos.long())
neg_emb = self.embedding_item(neg.long())
pos_scores= torch.sum(users_emb*pos_emb, dim=1)
neg_scores= torch.sum(users_emb*neg_emb, dim=1)
loss = torch.mean(nn.functional.softplus(neg_scores - pos_scores))
reg_loss = (1/2)*(users_emb.norm(2).pow(2) +
pos_emb.norm(2).pow(2) +
neg_emb.norm(2).pow(2))/float(len(users))
return loss, reg_loss
def forward(self, users, items):
users = users.long()
items = items.long()
users_emb = self.embedding_user(users)
items_emb = self.embedding_item(items)
scores = torch.sum(users_emb*items_emb, dim=1)
return self.f(scores)
class LightGCN(BasicModel):
def __init__(self,
config:dict,
dataset:BasicDataset):
super(LightGCN, self).__init__()
self.config = config
self.dataset : dataloader.BasicDataset = dataset
self.__init_weight()
def __init_weight(self):
self.num_users = self.dataset.n_users
self.num_items = self.dataset.m_items
self.latent_dim = self.config['latent_dim_rec']
self.n_layers = self.config['lightGCN_n_layers']
self.keep_prob = self.config['keep_prob']
self.A_split = self.config['A_split']
self.embedding_user = torch.nn.Embedding(
num_embeddings=self.num_users, embedding_dim=self.latent_dim)
self.embedding_item = torch.nn.Embedding(
num_embeddings=self.num_items, embedding_dim=self.latent_dim)
if self.config['pretrain'] == 0:
nn.init.normal_(self.embedding_user.weight, std=0.1)
nn.init.normal_(self.embedding_item.weight, std=0.1)
world.cprint('use NORMAL distribution initilizer')
else:
self.embedding_user.weight.data.copy_(torch.from_numpy(self.config['user_emb']))
self.embedding_item.weight.data.copy_(torch.from_numpy(self.config['item_emb']))
print('use pretarined data')
self.f = nn.Sigmoid()
self.Graph = self.dataset.getSparseGraph()
print(f"lgn is already to go(dropout:{self.config['dropout']})")
# print("save_txt")
def __dropout_x(self, x, keep_prob):
size = x.size()
index = x.indices().t()
values = x.values()
random_index = torch.rand(len(values)) + keep_prob
random_index = random_index.int().bool()
index = index[random_index]
values = values[random_index]/keep_prob
g = torch.sparse.FloatTensor(index.t(), values, size)
return g
def __dropout(self, keep_prob):
if self.A_split:
graph = []
for g in self.Graph:
graph.append(self.__dropout_x(g, keep_prob))
else:
graph = self.__dropout_x(self.Graph, keep_prob)
return graph
def computer(self):
"""
propagate methods for lightGCN
"""
users_emb = self.embedding_user.weight
items_emb = self.embedding_item.weight
all_emb = torch.cat([users_emb, items_emb])
# torch.split(all_emb , [self.num_users, self.num_items])
embs = [all_emb]
if self.config['dropout']:
if self.training:
print("droping")
g_droped = self.__dropout(self.keep_prob)
else:
g_droped = self.Graph
else:
g_droped = self.Graph
for layer in range(self.n_layers):
if self.A_split:
temp_emb = []
for f in range(len(g_droped)):
temp_emb.append(torch.sparse.mm(g_droped[f], all_emb))
side_emb = torch.cat(temp_emb, dim=0)
all_emb = side_emb
else:
all_emb = torch.sparse.mm(g_droped, all_emb)
embs.append(all_emb)
embs = torch.stack(embs, dim=1)
#print(embs.size())
light_out = torch.mean(embs, dim=1)
users, items = torch.split(light_out, [self.num_users, self.num_items])
return users, items
def getUsersRating(self, users):
all_users, all_items = self.computer()
users_emb = all_users[users.long()]
items_emb = all_items
rating = self.f(torch.matmul(users_emb, items_emb.t()))
return rating
def getEmbedding(self, users, pos_items, neg_items):
all_users, all_items = self.computer()
users_emb = all_users[users]
pos_emb = all_items[pos_items]
neg_emb = all_items[neg_items]
users_emb_ego = self.embedding_user(users)
pos_emb_ego = self.embedding_item(pos_items)
neg_emb_ego = self.embedding_item(neg_items)
return users_emb, pos_emb, neg_emb, users_emb_ego, pos_emb_ego, neg_emb_ego
def bpr_loss(self, users, pos, neg):
(users_emb, pos_emb, neg_emb,
userEmb0, posEmb0, negEmb0) = self.getEmbedding(users.long(), pos.long(), neg.long())
reg_loss = (1/2)*(userEmb0.norm(2).pow(2) +
posEmb0.norm(2).pow(2) +
negEmb0.norm(2).pow(2))/float(len(users))
pos_scores = torch.mul(users_emb, pos_emb)
pos_scores = torch.sum(pos_scores, dim=1)
neg_scores = torch.mul(users_emb, neg_emb)
neg_scores = torch.sum(neg_scores, dim=1)
loss = torch.mean(torch.nn.functional.softplus(neg_scores - pos_scores))
return loss, reg_loss
def forward(self, users, items):
# compute embedding
all_users, all_items = self.computer()
# print('forward')
#all_users, all_items = self.computer()
users_emb = all_users[users]
items_emb = all_items[items]
inner_pro = torch.mul(users_emb, items_emb)
gamma = torch.sum(inner_pro, dim=1)
return gamma
1.light Graph Convolution(LGC):
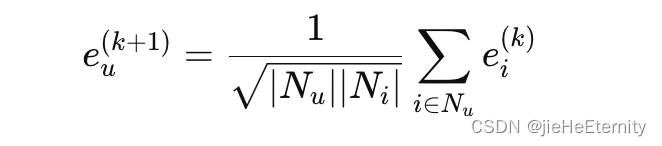

for layer in range(self.n_layers):
if self.A_split:
temp_emb = []
for f in range(len(g_droped)):
temp_emb.append(torch.sparse.mm(g_droped[f], all_emb))
side_emb = torch.cat(temp_emb, dim=0)
all_emb = side_emb
else:
all_emb = torch.sparse.mm(g_droped, all_emb)
embs.append(all_emb)
2.Layer Combination and Model Prediction:

embs = torch.stack(embs, dim=1)
light_out = torch.mean(embs, dim=1)
users, items = torch.split(light_out, [self.num_users, self.num_items])
3.Model Prediction:
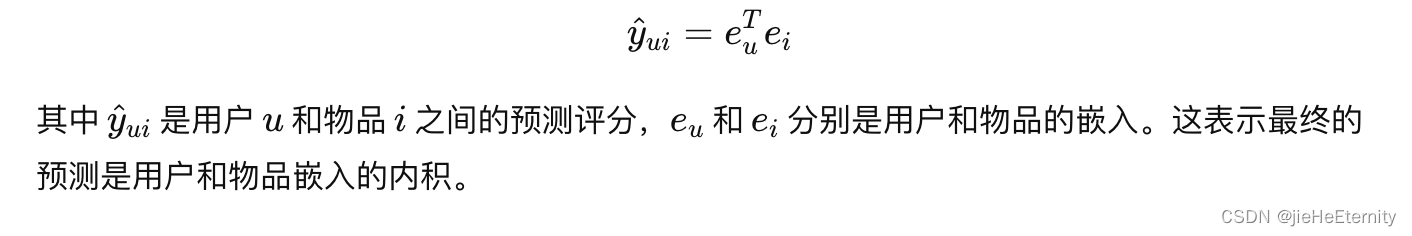
users_emb = all_users[users]
items_emb = all_items[items]
inner_pro = torch.mul(users_emb, items_emb)
gamma = torch.sum(inner_pro, dim=1)
return gamma
GSLRDA
from random import shuffle, choice
from util.relation import Relation
from util.config import Config
from util.io import FileIO
import tensorflow as tf
from util import config
import numpy as np
import scipy.sparse as sp
import random
class GSLRDA:
def __init__(self,conf,trainingSet=None,testSet=None):
self.config = conf
self.data = Relation(self.config, trainingSet, testSet)
self.num_ncRNAs, self.num_drugs, self.train_size = self.data.trainingSize()
self.emb_size = int(self.config['num.factors'])
self.maxIter = int(self.config['num.max.iter'])
learningRate = config.LineConfig(self.config['learnRate'])
self.lRate = float(learningRate['-init'])
regular = config.LineConfig(self.config['reg.lambda'])
self.regU, self.regI, self.regB = float(regular['-u']), float(regular['-i']), float(regular['-b'])
self.batch_size = int(self.config['batch_size'])
self.u_idx = tf.placeholder(tf.int32, name="u_idx")
self.v_idx = tf.placeholder(tf.int32, name="v_idx")
self.r = tf.placeholder(tf.float32, name="rating")
self.ncRNA_embeddings = tf.Variable(tf.truncated_normal(shape=[self.num_ncRNAs, self.emb_size], stddev=0.005),
name='U')
self.drug_embeddings = tf.Variable(tf.truncated_normal(shape=[self.num_drugs, self.emb_size], stddev=0.005),
name='V')
self.u_embedding = tf.nn.embedding_lookup(self.ncRNA_embeddings, self.u_idx)
self.v_embedding = tf.nn.embedding_lookup(self.drug_embeddings, self.v_idx)
config1 = tf.ConfigProto()
config1.gpu_options.allow_growth = True
self.sess = tf.Session(config=config1)
self.loss, self.lastLoss = 0, 0
args = config.LineConfig(self.config['SGL'])
self.ssl_reg = float(args['-lambda'])
self.drop_rate = float(args['-droprate'])
self.aug_type = int(args['-augtype'])
self.ssl_temp = float(args['-temp'])
norm_adj = self._create_adj_mat(is_subgraph=False)
norm_adj = self._convert_sp_mat_to_sp_tensor(norm_adj)
ego_embeddings = tf.concat([self.ncRNA_embeddings, self.drug_embeddings], axis=0)
s1_embeddings = ego_embeddings
s2_embeddings = ego_embeddings
all_s1_embeddings = [s1_embeddings]
all_s2_embeddings = [s2_embeddings]
all_embeddings = [ego_embeddings]
self.n_layers = 2
self._create_variable()
for k in range(0, self.n_layers):
if self.aug_type in [0, 1]:
self.sub_mat['sub_mat_1%d' % k] = tf.SparseTensor(
self.sub_mat['adj_indices_sub1'],
self.sub_mat['adj_values_sub1'],
self.sub_mat['adj_shape_sub1'])
self.sub_mat['sub_mat_2%d' % k] = tf.SparseTensor(
self.sub_mat['adj_indices_sub2'],
self.sub_mat['adj_values_sub2'],
self.sub_mat['adj_shape_sub2'])
else:
self.sub_mat['sub_mat_1%d' % k] = tf.SparseTensor(
self.sub_mat['adj_indices_sub1%d' % k],
self.sub_mat['adj_values_sub1%d' % k],
self.sub_mat['adj_shape_sub1%d' % k])
self.sub_mat['sub_mat_2%d' % k] = tf.SparseTensor(
self.sub_mat['adj_indices_sub2%d' % k],
self.sub_mat['adj_values_sub2%d' % k],
self.sub_mat['adj_shape_sub2%d' % k])
#s1 - view
for k in range(self.n_layers):
s1_embeddings = tf.sparse_tensor_dense_matmul(self.sub_mat['sub_mat_1%d' % k],s1_embeddings)
all_s1_embeddings += [s1_embeddings]
all_s1_embeddings = tf.stack(all_s1_embeddings, 1)
all_s1_embeddings = tf.reduce_mean(all_s1_embeddings, axis=1, keepdims=False)
self.s1_ncRNA_embeddings, self.s1_drug_embeddings = tf.split(all_s1_embeddings, [self.num_ncRNAs, self.num_drugs], 0)
#s2 - view
for k in range(self.n_layers):
s2_embeddings = tf.sparse_tensor_dense_matmul(self.sub_mat['sub_mat_2%d' % k],s2_embeddings)
all_s2_embeddings += [s2_embeddings]
all_s2_embeddings = tf.stack(all_s2_embeddings, 1)
all_s2_embeddings = tf.reduce_mean(all_s2_embeddings, axis=1, keepdims=False)
self.s2_ncRNA_embeddings, self.s2_drug_embeddings = tf.split(all_s2_embeddings, [self.num_ncRNAs, self.num_drugs], 0)
for k in range(self.n_layers):
ego_embeddings = tf.sparse_tensor_dense_matmul(norm_adj,ego_embeddings)
all_embeddings += [ego_embeddings]
all_embeddings = tf.stack(all_embeddings, 1)
all_embeddings = tf.reduce_mean(all_embeddings, axis=1, keepdims=False)
self.main_ncRNA_embeddings, self.main_drug_embeddings = tf.split(all_embeddings, [self.num_ncRNAs, self.num_drugs], 0)
self.neg_idx = tf.placeholder(tf.int32, name="neg_holder")
self.neg_drug_embedding = tf.nn.embedding_lookup(self.main_drug_embeddings, self.neg_idx)
self.u_embedding = tf.nn.embedding_lookup(self.main_ncRNA_embeddings, self.u_idx)
self.v_embedding = tf.nn.embedding_lookup(self.main_drug_embeddings, self.v_idx)
self.test = tf.reduce_sum(tf.multiply(self.u_embedding,self.main_drug_embeddings),1)
def _convert_sp_mat_to_sp_tensor(self, X):
coo = X.tocoo().astype(np.float32)
indices = np.mat([coo.row, coo.col]).transpose()
return tf.SparseTensor(indices, coo.data, coo.shape)
def _convert_csr_to_sparse_tensor_inputs(self, X):
coo = X.tocoo()
indices = np.mat([coo.row, coo.col]).transpose()
return indices, coo.data, coo.shape
def _create_variable(self):
self.sub_mat = {}
if self.aug_type in [0, 1]:
self.sub_mat['adj_values_sub1'] = tf.placeholder(tf.float32)
self.sub_mat['adj_indices_sub1'] = tf.placeholder(tf.int64)
self.sub_mat['adj_shape_sub1'] = tf.placeholder(tf.int64)
self.sub_mat['adj_values_sub2'] = tf.placeholder(tf.float32)
self.sub_mat['adj_indices_sub2'] = tf.placeholder(tf.int64)
self.sub_mat['adj_shape_sub2'] = tf.placeholder(tf.int64)
else:
for k in range(self.n_layers):
self.sub_mat['adj_values_sub1%d' % k] = tf.placeholder(tf.float32, name='adj_values_sub1%d' % k)
self.sub_mat['adj_indices_sub1%d' % k] = tf.placeholder(tf.int64, name='adj_indices_sub1%d' % k)
self.sub_mat['adj_shape_sub1%d' % k] = tf.placeholder(tf.int64, name='adj_shape_sub1%d' % k)
self.sub_mat['adj_values_sub2%d' % k] = tf.placeholder(tf.float32, name='adj_values_sub2%d' % k)
self.sub_mat['adj_indices_sub2%d' % k] = tf.placeholder(tf.int64, name='adj_indices_sub2%d' % k)
self.sub_mat['adj_shape_sub2%d' % k] = tf.placeholder(tf.int64, name='adj_shape_sub2%d' % k)
def _create_adj_mat(self, is_subgraph=False, aug_type=0):
n_nodes = self.num_ncRNAs + self.num_drugs
row_idx = [self.data.ncRNA[pair[0]] for pair in self.data.trainingData]
col_idx = [self.data.drug[pair[1]] for pair in self.data.trainingData]
if is_subgraph and aug_type in [0, 1, 2] and self.drop_rate > 0:
# data augmentation type --- 0: Node Dropout; 1: Edge Dropout; 2: Random Walk
if aug_type == 0:
drop_ncRNA_idx = random.sample(list(range(self.num_ncRNAs)), int(self.num_ncRNAs * self.drop_rate))
drop_drug_idx = random.sample(list(range(self.num_drugs)), int(self.num_drugs * self.drop_rate))
indicator_ncRNA = np.ones(self.num_ncRNAs, dtype=np.float32)
indicator_drug = np.ones(self.num_drugs, dtype=np.float32)
indicator_ncRNA[drop_ncRNA_idx] = 0.
indicator_drug[drop_drug_idx] = 0.
diag_indicator_ncRNA = sp.diags(indicator_ncRNA)
diag_indicator_drug = sp.diags(indicator_drug)
R = sp.csr_matrix(
(np.ones_like(row_idx, dtype=np.float32), (row_idx, col_idx)),
shape=(self.num_ncRNAs, self.num_drugs))
R_prime = diag_indicator_ncRNA.dot(R).dot(diag_indicator_drug)
(ncRNA_np_keep, drug_np_keep) = R_prime.nonzero()
ratings_keep = R_prime.data
tmp_adj = sp.csr_matrix((ratings_keep, (ncRNA_np_keep, drug_np_keep+self.num_ncRNAs)), shape=(n_nodes, n_nodes))
if aug_type in [1, 2]:
keep_idx = random.sample(list(range(self.data.elemCount())), int(self.data.elemCount() * (1 - self.drop_rate)))
ncRNA_np = np.array(row_idx)[keep_idx]
drug_np = np.array(col_idx)[keep_idx]
ratings = np.ones_like(ncRNA_np, dtype=np.float32)
tmp_adj = sp.csr_matrix((ratings, (ncRNA_np, drug_np+self.num_ncRNAs)), shape=(n_nodes, n_nodes))
else:
ncRNA_np = np.array(row_idx)
drug_np = np.array(col_idx)
ratings = np.ones_like(ncRNA_np, dtype=np.float32)
tmp_adj = sp.csr_matrix((ratings, (ncRNA_np, drug_np+self.num_ncRNAs)), shape=(n_nodes, n_nodes))
adj_mat = tmp_adj + tmp_adj.T
rowsum = np.array(adj_mat.sum(1))
d_inv = np.power(rowsum, -0.5).flatten()
d_inv[np.isinf(d_inv)] = 0.
d_mat_inv = sp.diags(d_inv)
norm_adj_tmp = d_mat_inv.dot(adj_mat)
adj_matrix = norm_adj_tmp.dot(d_mat_inv)
return adj_matrix
def calc_ssl_loss(self):
ncRNA_emb1 = tf.nn.embedding_lookup(self.s1_ncRNA_embeddings, tf.unique(self.u_idx)[0])
ncRNA_emb2 = tf.nn.embedding_lookup(self.s2_ncRNA_embeddings, tf.unique(self.u_idx)[0])
normalize_ncRNA_emb1 = tf.nn.l2_normalize(ncRNA_emb1, 1)
normalize_ncRNA_emb2 = tf.nn.l2_normalize(ncRNA_emb2, 1)
drug_emb1 = tf.nn.embedding_lookup(self.s1_drug_embeddings, tf.unique(self.v_idx)[0])
drug_emb2 = tf.nn.embedding_lookup(self.s2_drug_embeddings, tf.unique(self.v_idx)[0])
normalize_drug_emb1 = tf.nn.l2_normalize(drug_emb1, 1)
normalize_drug_emb2 = tf.nn.l2_normalize(drug_emb2, 1)
normalize_ncRNA_emb2_neg = normalize_ncRNA_emb2
normalize_drug_emb2_neg = normalize_drug_emb2
pos_score_ncRNA = tf.reduce_sum(tf.multiply(normalize_ncRNA_emb1, normalize_ncRNA_emb2), axis=1)
ttl_score_ncRNA = tf.matmul(normalize_ncRNA_emb1, normalize_ncRNA_emb2_neg, transpose_a=False, transpose_b=True)
pos_score_drug = tf.reduce_sum(tf.multiply(normalize_drug_emb1, normalize_drug_emb2), axis=1)
ttl_score_drug = tf.matmul(normalize_drug_emb1, normalize_drug_emb2_neg, transpose_a=False, transpose_b=True)
pos_score_ncRNA = tf.exp(pos_score_ncRNA / self.ssl_temp)
ttl_score_ncRNA = tf.reduce_sum(tf.exp(ttl_score_ncRNA / self.ssl_temp), axis=1)
pos_score_drug = tf.exp(pos_score_drug / self.ssl_temp)
ttl_score_drug = tf.reduce_sum(tf.exp(ttl_score_drug / self.ssl_temp), axis=1)
ssl_loss_ncRNA = -tf.reduce_sum(tf.log(pos_score_ncRNA / ttl_score_ncRNA)+1e-8)
ssl_loss_drug = -tf.reduce_sum(tf.log(pos_score_drug / ttl_score_drug)+1e-8)
ssl_loss = self.ssl_reg*(ssl_loss_ncRNA + ssl_loss_drug)
return ssl_loss
def calc_ssl_loss_v3(self):
ncRNA_emb1 = tf.nn.embedding_lookup(self.s1_ncRNA_embeddings, tf.unique(self.u_idx)[0])
ncRNA_emb2 = tf.nn.embedding_lookup(self.s2_ncRNA_embeddings, tf.unique(self.u_idx)[0])
drug_emb1 = tf.nn.embedding_lookup(self.s1_drug_embeddings, tf.unique(self.v_idx)[0])
drug_emb2 = tf.nn.embedding_lookup(self.s2_drug_embeddings, tf.unique(self.v_idx)[0])
emb_merge1 = tf.concat([ncRNA_emb1, drug_emb1], axis=0)
emb_merge2 = tf.concat([ncRNA_emb2, drug_emb2], axis=0)
normalize_emb_merge1 = tf.nn.l2_normalize(emb_merge1, 1)
normalize_emb_merge2 = tf.nn.l2_normalize(emb_merge2, 1)
pos_score = tf.reduce_sum(tf.multiply(normalize_emb_merge1, normalize_emb_merge2), axis=1)
ttl_score = tf.matmul(normalize_emb_merge1, normalize_emb_merge2, transpose_a=False, transpose_b=True)
pos_score = tf.exp(pos_score / self.ssl_temp)
ttl_score = tf.reduce_sum(tf.exp(ttl_score / self.ssl_temp), axis=1)
ssl_loss = -tf.reduce_sum(tf.log(pos_score / ttl_score))
ssl_loss = self.ssl_reg * ssl_loss
return ssl_loss
def next_batch_pairwise(self):
shuffle(self.data.trainingData)
batch_id = 0
while batch_id < self.train_size:
if batch_id + self.batch_size <= self.train_size:
ncRNAs = [self.data.trainingData[idx][0] for idx in range(batch_id, self.batch_size + batch_id)]
drugs = [self.data.trainingData[idx][1] for idx in range(batch_id, self.batch_size + batch_id)]
batch_id += self.batch_size
else:
ncRNAs = [self.data.trainingData[idx][0] for idx in range(batch_id, self.train_size)]
drugs = [self.data.trainingData[idx][1] for idx in range(batch_id, self.train_size)]
batch_id = self.train_size
u_idx, i_idx, j_idx = [], [], []
drug_list = list(self.data.drug.keys())
for i, ncRNA in enumerate(ncRNAs):
i_idx.append(self.data.drug[drugs[i]])
u_idx.append(self.data.ncRNA[ncRNA])
neg_drug = choice(drug_list)
while neg_drug in self.data.trainSet_u[ncRNA]:
neg_drug = choice(drug_list)
j_idx.append(self.data.drug[neg_drug])
yield u_idx, i_idx, j_idx
def buildModel(self):
y = tf.reduce_sum(tf.multiply(self.u_embedding, self.v_embedding), 1) \
- tf.reduce_sum(tf.multiply(self.u_embedding, self.neg_drug_embedding), 1)
rec_loss = -tf.reduce_sum(tf.log(tf.sigmoid(y))) + self.regU * (tf.nn.l2_loss(self.u_embedding) +
tf.nn.l2_loss(self.v_embedding) +
tf.nn.l2_loss(self.neg_drug_embedding))
ssl_loss = self.calc_ssl_loss_v3()
total_loss = rec_loss+ssl_loss
opt = tf.train.AdamOptimizer(self.lRate)
train = opt.minimize(total_loss)
init = tf.global_variables_initializer()
self.sess.run(init)
for iteration in range(self.maxIter):
sub_mat = {}
if self.aug_type in [0, 1]:
sub_mat['adj_indices_sub1'], sub_mat['adj_values_sub1'], sub_mat[
'adj_shape_sub1'] = self._convert_csr_to_sparse_tensor_inputs(
self._create_adj_mat(is_subgraph=True, aug_type=self.aug_type))
sub_mat['adj_indices_sub2'], sub_mat['adj_values_sub2'], sub_mat[
'adj_shape_sub2'] = self._convert_csr_to_sparse_tensor_inputs(
self._create_adj_mat(is_subgraph=True, aug_type=self.aug_type))
else:
for k in range(1, self.n_layers + 1):
sub_mat['adj_indices_sub1%d' % k], sub_mat['adj_values_sub1%d' % k], sub_mat[
'adj_shape_sub1%d' % k] = self._convert_csr_to_sparse_tensor_inputs(
self._create_adj_mat(is_subgraph=True, aug_type=self.aug_type))
sub_mat['adj_indices_sub2%d' % k], sub_mat['adj_values_sub2%d' % k], sub_mat[
'adj_shape_sub2%d' % k] = self._convert_csr_to_sparse_tensor_inputs(
self._create_adj_mat(is_subgraph=True, aug_type=self.aug_type))
for n, batch in enumerate(self.next_batch_pairwise()):
ncRNA_idx, i_idx, j_idx = batch
feed_dict = {self.u_idx: ncRNA_idx,
self.v_idx: i_idx,
self.neg_idx: j_idx, }
if self.aug_type in [0, 1]:
feed_dict.update({
self.sub_mat['adj_values_sub1']: sub_mat['adj_values_sub1'],
self.sub_mat['adj_indices_sub1']: sub_mat['adj_indices_sub1'],
self.sub_mat['adj_shape_sub1']: sub_mat['adj_shape_sub1'],
self.sub_mat['adj_values_sub2']: sub_mat['adj_values_sub2'],
self.sub_mat['adj_indices_sub2']: sub_mat['adj_indices_sub2'],
self.sub_mat['adj_shape_sub2']: sub_mat['adj_shape_sub2']
})
else:
for k in range(self.n_layers):
feed_dict.update({
self.sub_mat['adj_values_sub1%d' % k]: sub_mat['adj_values_sub1%d' % k],
self.sub_mat['adj_indices_sub1%d' % k]: sub_mat['adj_indices_sub1%d' % k],
self.sub_mat['adj_shape_sub1%d' % k]: sub_mat['adj_shape_sub1%d' % k],
self.sub_mat['adj_values_sub2%d' % k]: sub_mat['adj_values_sub2%d' % k],
self.sub_mat['adj_indices_sub2%d' % k]: sub_mat['adj_indices_sub2%d' % k],
self.sub_mat['adj_shape_sub2%d' % k]: sub_mat['adj_shape_sub2%d' % k]
})
_, l,rec_l,ssl_l = self.sess.run([train, total_loss, rec_loss, ssl_loss],feed_dict=feed_dict)
print('training:', iteration + 1, 'batch', n, 'rec_loss:', rec_l, 'ssl_loss',ssl_l)
if __name__ == '__main__':
conf = Config('GSLRDA.conf')
for i in range(0, 1):
train_path = f"./dataset/rtrain_{i}.txt"
test_path = f"./dataset/rtest_{i}.txt"
trainingData = FileIO.loadDataSet(conf, train_path, binarized=False, threshold=0)
testData = FileIO.loadDataSet(conf, test_path, bTest=True, binarized=False,
threshold=0)
re = GSLRDA(conf, trainingData, testData)
re.buildModel()
GSLRDA模型在论文中提出了几个主要的创新点,这些创新点在提供的代码中得到了实现。以下是论文中的创新点及其在代码中的对应部分:
整合主任务和辅助任务的多任务学习策略:
创新点:论文提出结合ncRNA与药物间的关联预测(主任务)和自监督学习(辅助任务)的多任务学习策略。
代码实现:
def buildModel(self):
# ...[代码省略]...
rec_loss = -tf.reduce_sum(tf.log(tf.sigmoid(y))) + self.regU * (tf.nn.l2_loss(self.u_embedding) +
tf.nn.l2_loss(self.v_embedding) +
tf.nn.l2_loss(self.neg_drug_embedding))
ssl_loss = self.calc_ssl_loss_v3()
total_loss = rec_loss + ssl_loss
# ...[代码省略]...
使用lightGCN学习ncRNA和药物的向量表示:
创新点:应用lightGCN框架来学习ncRNA和药物的嵌入。
代码实现:
for k in range(self.n_layers):
ego_embeddings = tf.sparse_tensor_dense_matmul(norm_adj, ego_embeddings)
all_embeddings += [ego_embeddings]
自监督学习的使用
创新点:通过数据增强生成不同视图,并进行比较学习来增强ncRNA和药物节点的表示。
def calc_ssl_loss_v3(self):
# ...[代码省略]...
pos_score = tf.exp(pos_score / self.ssl_temp)
ttl_score = tf.reduce_sum(tf.exp(ttl_score / self.ssl_temp), axis=1)
ssl_loss = -tf.reduce_sum(tf.log(pos_score / ttl_score))
ssl_loss = self.ssl_reg * ssl_loss
return ssl_loss
数据增强
创新点:使用节点dropout、边dropout和随机游走等方法生成ncRNA和药物节点的不同视图。
def _create_adj_mat(self, is_subgraph=False, aug_type=0):
# ...[代码省略]...
if aug_type == 0:
# Node Dropout
# ...[代码省略]...
if aug_type in [1, 2]:
# Edge Dropout or Random Walk
# ...[代码省略]...
# ...[代码省略]...
新颖的损失函数:
创新点:提出结合lightGCN损失和SSL损失的新颖损失函数。
def buildModel(self):
# ...[代码省略]...
total_loss = rec_loss + ssl_loss
# ...[代码省略]...
论文中对比学习的损失函数如下设计:


论文GSLRDA使用lightgcn学习向量的嵌入表示,使用数据增强和对比学习提升嵌入质量。
-
数据增强提供多样性:
数据增强通过改变原始数据的形式(如节点dropout、边dropout、随机游走等)来生成数据的不同视图。这些不同的视图提供了额外的信息和多样性,有助于模型学习更泛化的特征表示。
在图结构数据中,例如通过移除某些节点或边,可以模拟实际应用中可能遇到的不完整或嘈杂的数据情况,使模型更鲁棒。 -
对比学习强化区分能力:
对比学习通过鼓励模型将相似(正样本对)的样本靠近,将不相似(负样本对)的样本远离,来学习区分不同样本的能力。
这种学习方式使模型能够更好地理解数据中的细微差别,并通过这些差别来学习区分性强的特征。这对于学习具有区分力的嵌入表示至关重要。 -
结合主任务和辅助任务:
在自监督学习框架中,对比学习作为一个辅助任务,辅助主任务(例如分类或预测任务)的学习。这种结合可以促进模型在主任务上的性能,因为通过对比学习获得的更强区分能力和更泛化的特征表示可以直接应用于主任务。 -
提高泛化能力:
通过处理多个数据视图和学习从正负样本对中区分的能力,模型能够在面对新的、未见过的数据时表现出更好的泛化能力。这对于现实世界应用尤为重要,因为真实世界的数据往往更加多样化和复杂。
综上所述,数据增强和对比学习通过提供数据多样性和强化区分能力,共同作用于嵌入质量的提升,使得模型在主任务上的性能得到优化。
隐式反馈数据是指用户的行为数据,这些数据间接表达了用户的偏好或兴趣,而不是用户直接给出的明确反馈。在推荐系统中,隐式反馈数据与显式反馈数据相对,后者通常是用户直接给出的评分或评价。以下是一些常见的隐式反馈数据示例。

隐式反馈数据一般具有下面的特点:
非主动:用户没有明确表达喜欢或不喜欢,数据是通过用户的行为间接获得的。
正面偏差:隐式反馈数据通常只包含正面的信号(例如用户点击或购买了某商品),很少有直接的负面反馈(例如用户不喜欢某商品)。
噪声较多:隐式反馈数据可能包含很多不相关或误导性的信息,因为用户的行为不总是直接反映其真实的偏好。
在处理隐式反馈数据时,推荐系统的目标通常是从这些间接的、非主动的行为中推断出用户的潜在兴趣和偏好。这与处理显式反馈数据(如用户评分)不同,后者用户直接表达了对项目的喜好程度。隐式反馈的处理通常更具挑战性,但也更贴近真实世界的应用场景。
使用NGCF模型处理(用户,项目,0或者1)这样的交互数据时的流程。