文章目录
- 简介
- 安装
- ```datasets download```下载基因组/基因序列
- 按照GCA list文件编号下载
- 下载大基因组
- genome完整参数
- gene参数
- ```datasets summary```下载元数据
- ```dataformat```将json转换成表格格式
- 通过json文件解析其他字段
- 问题
简介
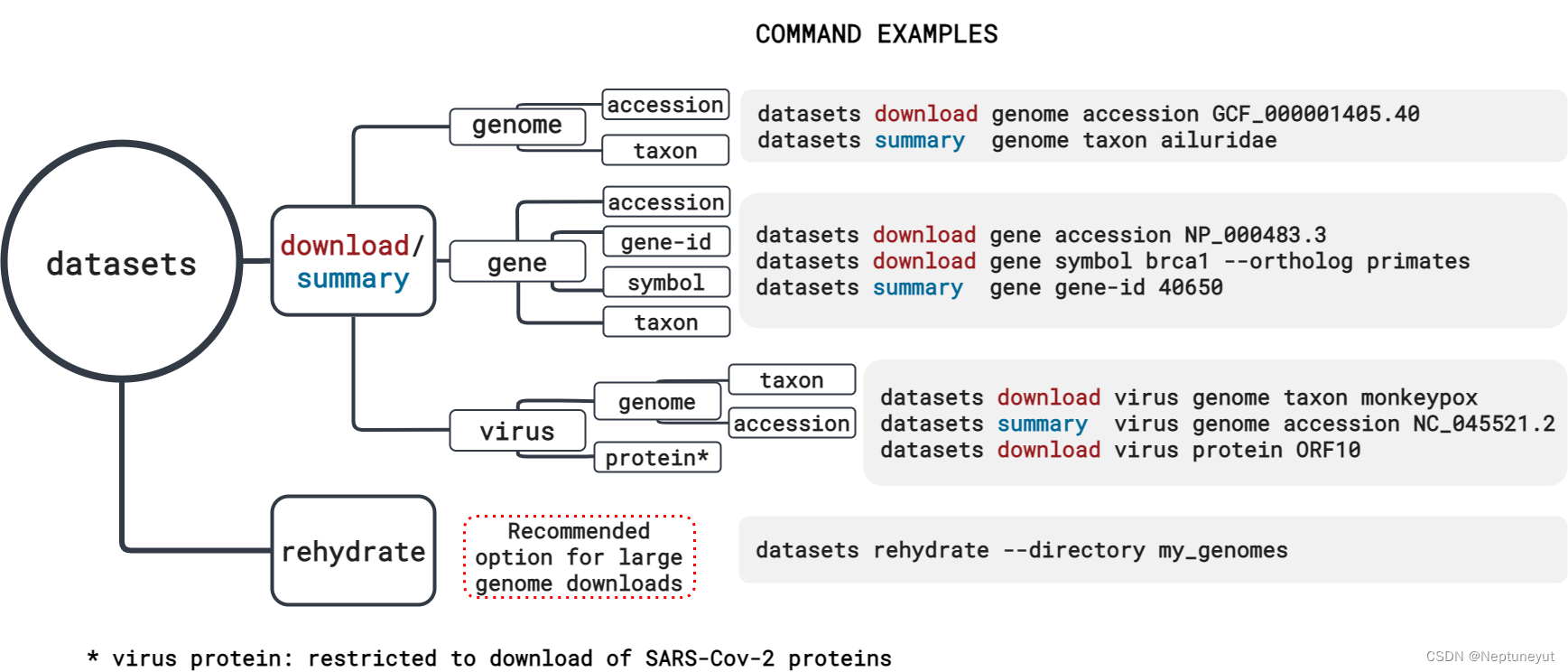
NCBI Datasets 可以轻松从 NCBI 数据库中收集数据。使用命令行界面(CLI)工具或 NCBI Datasets 网页界面查找和下载基因和基因组的序列、注释和元数据。如下是可用的工具:
安装
- 使用conda安装Datasets CLI tools,
datasetsand dataformat:
# 注意不是datasets而是ncbi-datasets-cli
$ conda install -c conda-forge ncbi-datasets-cli
(base) [yut@io02 ~]$ datasets --version
datasets version: 15.25.0
datasets download下载基因组/基因序列
datasets从 NCBI 下载所有生命领域的生物序列数据,dataformat将前者下载的数据包中的元数据从 JSON Lines 格式转换为其他格式。
使用datasets下载人类参考基因组 GRCh38 的基因组数据包:
$ datasets download genome taxon human --reference --filename human-reference.zip
使用 dataformat从下载的人类参考基因组 GRCh38 数据包中提取选定的元数据字段:
$ dataformat tsv genome --package human-reference.zip --fields organism-name,assminfo-name,accession,assminfo-submitter
Organism name Assembly Name Assembly Accession Assembly Submitter
Homo sapiens GRCh38.p14 GCF_000001405.40 Genome Reference Consortium
按照GCA list文件编号下载
(base) [yut@io02 02_Glacier_new_taxa]$ head 3.gca
GCF_020042285.1
GCF_020783315.1
GCF_024343615.1
(base) [yut@io02 02_Glacier_new_taxa]$ time datasets download genome accession --inputfile 3.gca --include gff3,rna,cds,protein,genome,seq-report --filename 3genome.zip
# --inputfile:输入GCA号的list,每行一个
# --filename:输出zip包名称,默认ncbi-dataset.zip
New version of client (15.27.1) available at https://ftp.ncbi.nlm.nih.gov/pub/datasets/command-line/LATEST/linux-amd64/datasets
Collecting 3 genome records [================================================] 100% 3/3
Downloading: 3genome.zip 10.4MB valid zip archive
Validating package files [================================================] 100% 18/18
real 0m8.208s
user 0m0.652s
sys 0m0.234s
(base) [yut@io02 02_Glacier_new_taxa]$ ls
3genome.zip download.log
下载大基因组
下载大量基因组,首先下载压缩包,然后分三步访问数据。
- 1.下载人基因组压缩包
datasets download genome accession GCF_000001405.40 --dehydrated --filename human_GRCh38_dataset.zip
- 2.解压
unzip human_GRCh38_dataset.zip -d my_human_dataset
- 3.转换格式
datasets rehydrate --directory my_human_dataset/
genome完整参数
(base) [yut@io02 ~]$ datasets download genome --help
Download a genome data package. Genome data packages may include genome, transcript and protein sequences, annotation and one or more data reports. Data packages are downloaded as a zip archive.
The default genome data package includes the following files:
* <accession>_<assembly_name>_genomic.fna (genomic sequences)
* assembly_data_report.jsonl (data report with genome assembly and annotation metadata)
* dataset_catalog.json (a list of files and file types included in the data package)
Usage
datasets download genome [flags]
datasets download genome [command]
Sample Commands
datasets download genome accession GCF_000001405.40 --chromosomes X,Y --include genome,gff3,rna
datasets download genome taxon "bos taurus" --dehydrated
datasets download genome taxon human --assembly-level chromosome,complete --dehydrated
datasets download genome taxon mouse --search C57BL/6J --search "Broad Institute" --dehydrated
Available Commands
accession Download a genome data package by Assembly or BioProject accession
taxon Download a genome data package by taxon (NCBI Taxonomy ID, scientific or common name at any tax rank)
Flags
--annotated Limit to annotated genomes
--assembly-level string Limit to genomes at one or more assembly levels (comma-separated):
* chromosome
* complete
* contig
* scaffold
(default "[]")
--assembly-source string Limit to 'RefSeq' (GCF_) or 'GenBank' (GCA_) genomes (default "all")
--chromosomes strings Limit to a specified, comma-delimited list of chromosomes, or 'all' for all chromosomes
--dehydrated Download a dehydrated zip archive including the data report and locations of data files (use the rehydrate command to retrieve data files).
--exclude-atypical Exclude atypical assemblies
--mag string Limit to metagenome assembled genomes (only) or remove them from the results (exclude) (default "all")
--preview Show information about the requested data package
--reference Limit to reference genomes
--released-after string Limit to genomes released on or after a specified date (MM/DD/YYYY)
--released-before string Limit to genomes released on or before a specified date (MM/DD/YYYY)
--search strings Limit results to genomes with specified text in the searchable fields:
species and infraspecies, assembly name and submitter.
To search multiple strings, use the flag multiple times.
Global Flags
--api-key string Specify an NCBI API key
--debug Emit debugging info
--filename string Specify a custom file name for the downloaded data package (default "ncbi_dataset.zip")
--help Print detailed help about a datasets command
--no-progressbar Hide progress bar
--version Print version of datasets
Use datasets download genome <command> --help for detailed help about a command.
gene参数
(base) [yut@io02 ~]$ datasets download gene --help
Download a gene data package. Gene data packages include gene, transcript and protein sequences and one or more data reports. Data packages are downloaded as a zip archive.
The default gene data package for NM, NR, NP, XM, XR, XP and YP accessions:
* rna.fna (transcript sequences)
* protein.faa (protein sequences)
* data_report.jsonl (data report with gene metadata)
* dataset_catalog.json (a list of files and file types included in the data package)
Usage
datasets download gene [flags]
datasets download gene [command]
Sample Commands
datasets download gene gene-id 672
datasets download gene symbol brca1 --taxon mouse
datasets download gene accession NP_000483.3
datasets download gene gene-id 2778 --fasta-filter NC_000020.11,NM_001077490.3,NP_001070958.1
Available Commands
gene-id Download a gene data package by NCBI Gene ID
symbol Download a gene data package by gene symbol
accession Download a gene data package by RefSeq nucleotide or protein accession
taxon Download a gene data package by taxon (NCBI Taxonomy ID, scientific or common name at any tax rank)
Flags
--fasta-filter strings Limit protein and RNA sequence files to the specified RefSeq nucleotide and protein accessions
--fasta-filter-file string Limit protein and RNA sequence files to the specified RefSeq nucleotide and protein accessions included in the specified file
--preview Show information about the requested data package
Global Flags
--api-key string Specify an NCBI API key
--debug Emit debugging info
--filename string Specify a custom file name for the downloaded data package (default "ncbi_dataset.zip")
--help Print detailed help about a datasets command
--no-progressbar Hide progress bar
--version Print version of datasets
Use datasets download gene <command> --help for detailed help about a command.
datasets summary下载元数据
(base) [yut@io02 ~]$ datasets summary --help
Print a data report containing gene, genome or virus metadata in JSON format.
Usage
datasets summary [flags]
datasets summary [command]
Sample Commands
datasets summary genome accession GCF_000001405.40
datasets summary genome taxon "mus musculus"
datasets summary gene gene-id 672
datasets summary gene symbol brca1 --taxon mouse
datasets summary gene accession NP_000483.3
datasets summary virus genome accession NC_045512.2
datasets summary virus genome taxon sars-cov-2 --host dog
Available Commands
gene Print a summary of a gene dataset
genome Print a data report containing genome metadata
virus Print a data report containing virus genome metadata
Global Flags
--api-key string Specify an NCBI API key
--debug Emit debugging info
--help Print detailed help about a datasets command
--version Print version of datasets
Use datasets summary <command> --help for detailed help about a command.
- 实例
(base) [yut@io02 ~]$ datasets summary genome accession GCF_000001405.40
New version of client (15.27.1) available at https://ftp.ncbi.nlm.nih.gov/pub/datasets/command-line/LATEST/linux-amd64/datasets
{"reports": [{"accession":"GCF_000001405.40","annotation_info":{"busco":{"busco_lineage":"primates_odb10","busco_ver":"4.1.4","complete":0.99187225,"duplicated":0.007256894,"fragmented":0.0015239477,"missing":0.0066037737,"single_copy":0.9846154,"total_count":"13780"},"method":"Best-placed RefSeq; Gnomon; RefSeqFE; cmsearch; tRNAscan-SE","name":"GCF_000001405.40-RS_2023_10","pipeline":"NCBI eukaryotic genome annotation pipeline","provider":"NCBI RefSeq","release_date":"2023-10-02","report_url":"https://www.ncbi.nlm.nih.gov/genome/annotation_euk/Homo_sapiens/GCF_000001405.40-RS_2023_10.html","software_version":"10.2","stats":{"gene_counts":{"non_coding":22158,"other":413,"protein_coding":20080,"pseudogene":17001,"total":59652}},"status":"Updated annotation"},"assembly_info":{"assembly_level":"Chromosome","assembly_name":"GRCh38.p14","assembly_status":"current","assembly_type":"haploid-with-alt-loci","bioproject_accession":"PRJNA31257","bioproject_lineage":[{"bioprojects":[{"accession":"PRJNA31257","title":"The Human Genome Project, currently maintained by the Genome Reference Consortium (GRC)"}]}],"blast_url":"https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch\u0026PROG_DEF=blastn\u0026BLAST_SPEC=GDH_GCF_000001405.40","description":"Genome Reference Consortium Human Build 38 patch release 14 (GRCh38.p14)","paired_assembly":{"accession":"GCA_000001405.29","only_genbank":"4 unlocalized and unplaced scaffolds.","status":"current"},"refseq_category":"reference genome","release_date":"2022-02-03","submitter":"Genome Reference Consortium","synonym":"hg38"},"assembly_stats":{"contig_l50":18,"contig_n50":57879411,"gaps_between_scaffolds_count":349,"gc_count":"1374283647","gc_percent":41,"number_of_component_sequences":35611,"number_of_contigs":996,"number_of_organelles":1,"number_of_scaffolds":470,"scaffold_l50":16,"scaffold_n50":67794873,"total_number_of_chromosomes":24,"total_sequence_length":"3099441038","total_ungapped_length":"2948318359"},"current_accession":"GCF_000001405.40","organelle_info":[{"description":"Mitochondrion","submitter":"Genome Reference Consortium","total_seq_length":"16569"}],"organism":{"common_name":"human","organism_name":"Homo sapiens","tax_id":9606},"paired_accession":"GCA_000001405.29","source_database":"SOURCE_DATABASE_REFSEQ"}],"total_count": 1}
(base) [yut@io02 ~]$ datasets summary gene gene-id 672
New version of client (15.27.1) available at https://ftp.ncbi.nlm.nih.gov/pub/datasets/command-line/LATEST/linux-amd64/datasets
{"reports": [{"gene":{"annotations":[{"annotation_name":"GCF_000001405.40-RS_2023_10","annotation_release_date":"2023-10-02","assembly_accession":"GCF_000001405.40","assembly_name":"GRCh38.p14","genomic_locations":[{"genomic_accession_version":"NC_000017.11","genomic_range":{"begin":"43044295","end":"43170327","orientation":"minus"},"sequence_name":"17"}]},{"annotation_name":"GCF_009914755.1-RS_2023_10","annotation_release_date":"2023-10-02","assembly_accession":"GCF_009914755.1","assembly_name":"T2T-CHM13v2.0","genomic_locations":[{"genomic_accession_version":"NC_060941.1","genomic_range":{"begin":"43902857","end":"44029084","orientation":"minus"},"sequence_name":"17"}]}],"chromosomes":["17"],"common_name":"human","description":"BRCA1 DNA repair associated","ensembl_gene_ids":["ENSG00000012048"],"gene_groups":[{"id":"672","method":"NCBI Ortholog"}],"gene_id":"672","nomenclature_authority":{"authority":"HGNC","identifier":"HGNC:1100"},"omim_ids":["113705"],"orientation":"minus","protein_count":368,"reference_standards":[{"gene_range":{"accession_version":"NG_005905.2","range":[{"begin":"92501","end":"173689","orientation":"plus"}]},"type":"REFSEQ_GENE"}],"swiss_prot_accessions":["P38398"],"symbol":"BRCA1","synonyms":["IRIS","PSCP","BRCAI","BRCC1","FANCS","PNCA4","RNF53","BROVCA1","PPP1R53"],"tax_id":"9606","taxname":"Homo sapiens","transcript_count":368,"transcript_type_counts":[{"count":368,"type":"PROTEIN_CODING"}],"type":"PROTEIN_CODING"},"query":["672"]}],"total_count": 1}
- 下载结果为json格式
dataformat将json转换成表格格式
(base) [yut@io02 ~]$ dataformat tsv
Convert data to TSV format.
Refer to NCBI's [download and install](https://www.ncbi.nlm.nih.gov/datasets/docs/v2/download-and-install/) documentation for information about getting started with the command-line tools.
Usage
dataformat tsv [command]
Report Commands
genome Convert Genome Assembly Data Report into TSV format
genome-seq Convert Genome Assembly Sequence Report into TSV format
gene Convert Gene Report into TSV format
gene-product Convert Gene Product Report into TSV format
virus-genome Convert Virus Data Report into TSV format
virus-annotation Convert Virus Annotation Report into TSV format
microbigge Convert MicroBIGG-E Data Report into TSV format
prok-gene Convert Prokaryote Gene Report into TSV format
prok-gene-location Convert Prokaryote Gene Location Report into TSV format
genome-annotations Convert Genome Annotation Report into TSV format
Flags
--elide-header Do not output header
-h, --help help for tsv
Global Flags
--force Force dataformat to run without type check prompt
Use dataformat tsv <command> --help for detailed help about a command.
(base) [yut@io02 ~]$ dataformat tsv gene
Error: --inputfile and/or --packagefile must be specified, or data can be read from standard input
Usage
dataformat tsv gene [flags]
Examples
dataformat tsv gene --inputfile gene_package/ncbi_dataset/data/data_report.jsonl
dataformat tsv gene --package genes.zip
Flags
--fields strings Comma-separated list of fields (default annotation-assembly-accession,annotation-assembly-name,annotation-genomic-range-accession,annotation-genomic-range-exon-order,annotation-genomic-range-exon-orientation,annotation-genomic-range-exon-start,annotation-genomic-range-exon-stop,annotation-genomic-range-range-order,annotation-genomic-range-range-orientation,annotation-genomic-range-range-start,annotation-genomic-range-range-stop,annotation-genomic-range-seq-name,annotation-release-date,annotation-release-name,chromosomes,common-name,description,ensembl-geneids,gene-id,gene-type,genomic-region-gene-range-accession,genomic-region-gene-range-range-order,genomic-region-gene-range-range-orientation,genomic-region-gene-range-range-start,genomic-region-gene-range-range-stop,genomic-region-genomic-region-type,group-id,group-method,name-authority,name-id,omim-ids,orientation,protein-count,ref-standard-gene-range-accession,ref-standard-gene-range-range-order,ref-standard-gene-range-range-orientation,ref-standard-gene-range-range-start,ref-standard-gene-range-range-stop,ref-standard-genomic-region-type,replaced-gene-id,rna-type,swissprot-accessions,symbol,synonyms,tax-id,tax-name,transcript-count)
- annotation-assembly-accession
- annotation-assembly-name
- annotation-genomic-range-accession
- annotation-genomic-range-exon-order
- annotation-genomic-range-exon-orientation
- annotation-genomic-range-exon-start
- annotation-genomic-range-exon-stop
- annotation-genomic-range-range-order
- annotation-genomic-range-range-orientation
- annotation-genomic-range-range-start
- annotation-genomic-range-range-stop
- annotation-genomic-range-seq-name
- annotation-release-date
- annotation-release-name
- chromosomes
- common-name
- description
- ensembl-geneids
- gene-id
- gene-type
- genomic-region-gene-range-accession
- genomic-region-gene-range-range-order
- genomic-region-gene-range-range-orientation
- genomic-region-gene-range-range-start
- genomic-region-gene-range-range-stop
- genomic-region-genomic-region-type
- group-id
- group-method
- name-authority
- name-id
- omim-ids
- orientation
- protein-count
- ref-standard-gene-range-accession
- ref-standard-gene-range-range-order
- ref-standard-gene-range-range-orientation
- ref-standard-gene-range-range-start
- ref-standard-gene-range-range-stop
- ref-standard-genomic-region-type
- replaced-gene-id
- rna-type
- swissprot-accessions
- symbol
- synonyms
- tax-id
- tax-name
- transcript-count
-h, --help help for gene
--inputfile string Input file (default "ncbi_dataset/data/data_report.jsonl")
--package string Data package (zip archive), inputfile parameter is relative to the root path inside the archive
Global Flags
--elide-header Do not output header
--force Force dataformat to run without type check prompt
- 实例
(base) [yut@io02 ~]$ datasets summary gene gene-id 672 --as-json-lines |dataformat tsv gene
New version of client (15.27.1) available at https://ftp.ncbi.nlm.nih.gov/pub/datasets/command-line/LATEST/linux-amd64/datasets
Annotation Assembly Accession Annotation Assembly Name Annotation Genomic Range Accession Annotation Genomic Range Exons Order Annotation Genomic Range Exons Orientation Annotation Genomic Range Exons Start Annotation Genomic Range Exons Stop Annotation Genomic Range Order Annotation Genomic Range Orientation Annotation Genomic Range Start Annotation Genomic Range Stop Annotation Genomic Range Seq Name Annotation Release Date Annotation Release Name Chromosomes Common Name Description Ensembl GeneIDs NCBI GeneID Gene Type Genomic Region Gene Range Sequence Accession Genomic Region Gene Range Order Genomic Region Gene Range Orientation Genomic Region Gene Range Start Genomic Region Gene Range Stop Genomic Region Genomic Region Type Gene Group Identifier Gene Group Method Nomenclature Authority Nomenclature ID OMIM IDs Orientation Proteins Reference Standard Gene Range Sequence Accession Reference Standard Gene Range Order Reference Standard Gene Range Orientation Reference Standard Gene Range Start Reference Standard Gene Range Stop Reference Standard Genomic Region Type Replaced NCBI GeneID RNA Type SwissProt Accessions Symbol Synonyms Taxonomic ID Taxonomic Name Transcripts
GCF_000001405.40 GRCh38.p14 NC_000017.11 minus 43044295 43170327 17 2023-10-02 GCF_000001405.40-RS_2023_10 17 human BRCA1 DNA repair associated ENSG00000012048 672 PROTEIN_CODING 672 NCBI Ortholog HGNC HGNC:1100 113705 minus 368 NG_005905.2 plus 92501 173689 REFSEQ_GENE P38398 BRCA1 IRIS,PSCP,BRCAI,BRCC1,FANCS,PNCA4,RNF53,BROVCA1,PPP1R53 9606 Homo sapiens 368
GCF_009914755.1 T2T-CHM13v2.0 NC_060941.1 minus 43902857 44029084 17 2023-10-02 GCF_009914755.1-RS_2023_10 17 human BRCA1 DNA repair associated ENSG00000012048 672 PROTEIN_CODING 672 NCBI Ortholog HGNC HGNC:1100 113705 minus 368 NG_005905.2 plus 92501 173689 REFSEQ_GENE P38398 BRCA1 IRIS,PSCP,BRCAI,BRCC1,FANCS,PNCA4,RNF53,BROVCA1,PPP1R53 9606 Homo sapiens 368
(base) [yut@io02 ~]$ datasets summary gene gene-id 672 --as-json-lines |dataformat tsv gene --fields gene-id,gene-type,symbol
New version of client (15.27.1) available at https://ftp.ncbi.nlm.nih.gov/pub/datasets/command-line/LATEST/linux-amd64/datasets
NCBI GeneID Gene Type Symbol
672 PROTEIN_CODING BRCA1
# --as-json-lines必须加上
# --fields指定需要的字段,多个空格隔开
通过json文件解析其他字段
- 某些字段无法通过
dataformat提取出来,可先保存成json文件,然后通过下面脚本解析:
(base) [yut@node01 ~]$ cat dataset.json
{"accession":"GCA_013141435.1","annotation_info":{"method":"Best-placed reference protein set; GeneMarkS-2+","name":"NCBI Prokaryotic Genome Annotation Pipeline (PGAP)","pipeline":"NCBI Prokaryotic Genome Annotation Pipeline (PGAP)","provider":"NCBI","release_date":"2020-05-14","software_version":"4.11","stats":{"gene_counts":{"non_coding":27,"protein_coding":2566,"pseudogene":17,"total":2610}}},"assembly_info":{"assembly_level":"Contig","assembly_method":"MetaSPAdes v. 3.10.1","assembly_name":"ASM1314143v1","assembly_status":"current","assembly_type":"haploid","bioproject_accession":"PRJNA622654","bioproject_lineage":[{"bioprojects":[{"accession":"PRJNA622654","title":"Metagenomic profiling of ammonia and methane-oxidizing microorganisms in a Dutch drinking water treatment plant"}]}],"biosample":{"accession":"SAMN14539096","attributes":[{"name":"isolation_source","value":"Primary rapid sand filter"},{"name":"collection_date","value":"not applicable"},{"name":"geo_loc_name","value":"Netherlands"},{"name":"lat_lon","value":"not applicable"},{"name":"isolate","value":"P-RSF-IL-07"},{"name":"depth","value":"not applicable"},{"name":"env_broad_scale","value":"drinking water treatment plant"},{"name":"env_local_scale","value":"Primary rapid sand filter"},{"name":"env_medium","value":"not applicable"},{"name":"metagenomic","value":"1"},{"name":"environmental-sample","value":"1"},{"name":"sample_type","value":"metagenomic assembly"},{"name":"metagenome-source","value":"drinking water metagenome"},{"name":"derived_from","value":"This BioSample is a metagenomic assembly obtained from the drinking water metagenome BioSample:SAMN14524263, SAMN14524264, SAMN14524265, SAMN14524266"}],"bioprojects":[{"accession":"PRJNA622654"}],"description":{"comment":"Keywords: GSC:MIxS;MIMAG:6.0","organism":{"organism_name":"Ferruginibacter sp.","tax_id":1940288},"title":"MIMAG Metagenome-assembled Genome sample from Ferruginibacter sp."},"last_updated":"2020-05-19T00:50:12.857","models":["MIMAG.water"],"owner":{"contacts":[{}],"name":"Radboud University"},"package":"MIMAG.water.6.0","publication_date":"2020-05-19T00:50:12.857","sample_ids":[{"label":"Sample name","value":"Ferruginibacter sp. P-RSF-IL-07"}],"status":{"status":"live","when":"2020-05-19T00:50:12.857"},"submission_date":"2020-04-04T13:17:04.950"},"comments":"The annotation was added by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP). Information about PGAP can be found here: https://www.ncbi.nlm.nih.gov/genome/annotation_prok/","genome_notes":["derived from metagenome"],"release_date":"2020-05-21","sequencing_tech":"Illumina MiSeq","submitter":"Radboud University"},"assembly_stats":{"contig_l50":10,"contig_n50":104094,"gc_count":"978119","gc_percent":32,"genome_coverage":"270.4x","number_of_component_sequences":43,"number_of_contigs":43,"total_sequence_length":"3056910","total_ungapped_length":"3056910"},"average_nucleotide_identity":{"best_ani_match":{"ani":79.65,"assembly":"GCA_003426875.1","assembly_coverage":0.01,"category":"type","organism_name":"Lutibacter oceani","type_assembly_coverage":0.01},"category":"category_na","comment":"na","match_status":"low_coverage","submitted_organism":"Ferruginibacter sp.","submitted_species":"Ferruginibacter sp.","taxonomy_check_status":"Inconclusive"},"current_accession":"GCA_013141435.1","organism":{"infraspecific_names":{"isolate":"P-RSF-IL-07"},"organism_name":"Ferruginibacter sp.","tax_id":1940288},"source_database":"SOURCE_DATABASE_GENBANK","wgs_info":{"master_wgs_url":"https://www.ncbi.nlm.nih.gov/nuccore/JABFQZ000000000.1","wgs_contigs_url":"https://www.ncbi.nlm.nih.gov/Traces/wgs/JABFQZ01","wgs_project_accession":"JABFQZ01"}}
(base) [yut@node01 ~]$ Parse_dataset_genome_json_metadata.py *json
Save result in output.csv
(base) [yut@node01 ~]$ cat output.csv
Accession,Geo Location Name,Latitude and Longitude,Collection date,Env broad scale,Env local scale,Env medium,Sample type
GCA_013141435.1,Netherlands,not applicable,not applicable,drinking water treatment plant,Primary rapid sand filter,not applicable,metagenomic assembly
(base) [yut@node01 ~]$ cat ~/Software/Important_scripts/Parse_dataset_genome_json_metadata.py
#!/usr/bin/env python
import argparse
import json
import pandas as pd
# 创建参数解析器
parser = argparse.ArgumentParser(description='Parse JSON data')
parser.add_argument('json_file', help='Path to the JSON file')
# 解析参数
args = parser.parse_args()
# 读取JSON文件
with open(args.json_file, 'r') as file:
json_str = file.read()
# 解析JSON
data = json.loads(json_str)
# 获取env_broad_scale字段的值
# 获取所需字段的值
accession = data["accession"]
geo_loc_name = data["assembly_info"]["biosample"]["attributes"][2]["value"]
lat_lon = data["assembly_info"]["biosample"]["attributes"][3]["value"]
collection_date = data['assembly_info']['biosample']['attributes'][1]['value']
env_broad_scale = data["assembly_info"]["biosample"]["attributes"][6]["value"]
env_local_scale = data['assembly_info']['biosample']['attributes'][7]['value']
env_medium = data['assembly_info']['biosample']['attributes'][8]['value']
sample_type = data['assembly_info']['biosample']['attributes'][11]['value']
# output
# 创建DataFrame
df = pd.DataFrame({
'Accession': [accession],
'Geo Location Name': [geo_loc_name],
'Latitude and Longitude': [lat_lon],
'Collection date' : [collection_date],
'Env broad scale' : [env_broad_scale],
'Env local scale' : [env_local_scale],
'Env medium' : [env_medium],
'Sample type' : [sample_type]
})
# 将DataFrame保存为CSV文件
df.to_csv('output.csv', index=False)
print('Save result in output.csv ')
# run
$ (base) [yut@node01 ~]$ Parse_dataset_genome_json_metadata.py dataset.json
Save result in output.csv
# output
(base) [yut@node01 ~]$ cat output.csv
Accession,Geo Location Name,Latitude and Longitude,Collection date,Env broad scale,Env local scale,Env medium,Sample type
GCA_013141435.1,Netherlands,not applicable,not applicable,drinking water treatment plant,Primary rapid sand filter,not applicable,metagenomic assembly
问题
- Error: Internal error (invalid zip archive)并且没有输出文件
(base) [yut@io02 02_Glacier_new_taxa]$ time datasets download genome accession --inputfile GTDB_R214_63_Ferruginibacter_genus.GCA --include gff3,rna,cds,protein,genome,seq-report
New version of client (15.27.1) available at https://ftp.ncbi.nlm.nih.gov/pub/datasets/command-line/LATEST/linux-amd64/datasets
Collecting 63 genome records [================================================] 100% 63/63
Downloading: ncbi_dataset.zip 146MB done
Validating package files [===========>------------------------------------] 28% 70/252
Error: Internal error (invalid zip archive). Please try again
上述问题可能是输入编号既包括GCA又包括GCF编号,解决办法是将两者分开下载,或者等到Validating package files停掉命令