目录
一、实验介绍
1. 论文:基于多模态深度学习方法的单细胞多组学数据聚类
Abstract
2. Github链接
二、实验环境
0. 作者要求
1. 环境复现
实验一
实验二(本实验)
2. 库版本介绍
实验一
实验二
3. IDE
三、实验内容
1. 用法
2. 输出
3. 参数
4. run_scMDC
设置超参数
对scRNA-seq数据进行预处理
构建scMultiCluster模型
预训练
使用KMeans确定聚类数k
微调模型,训练聚类层
保存预测结果和embedding到文件
输出预测结果y_pred,计算指标AMI、NMI和ARI
5. 聚类结果可视化
可视化结果
附录:run_scMDC训练过程
一、实验介绍
1. 论文:基于多模态深度学习方法的单细胞多组学数据聚类
Clustering of single-cell multi-omics data with a multimodal deep learning method | Nature Communications![]() https://www.nature.com/articles/s41467-022-35031-9
https://www.nature.com/articles/s41467-022-35031-9
Abstract
Single-cell multimodal sequencing technologies are developed to simultaneously profile different modalities of data in the same cell. It provides a unique opportunity to jointly analyze multimodal data at the single-cell level for the identification of distinct cell types. A correct clustering result is essential for the downstream complex biological functional studies. However, combining different data sources for clustering analysis of single-cell multimodal data remains a statistical and computational challenge. Here, we develop a novel multimodal deep learning method, scMDC, for single-cell multi-omics data clustering analysis. scMDC is an end-to-end deep model that explicitly characterizes different data sources and jointly learns latent features of deep embedding for clustering analysis. Extensive simulation and real-data experiments reveal that scMDC outperforms existing single-cell single-modal and multimodal clustering methods on different single-cell multimodal datasets. The linear scalability of running time makes scMDC a promising method for analyzing large multimodal datasets.
开发单细胞多模态测序技术以同时分析同一细胞中的不同数据模式。它提供了一个独特的机会,可以在单细胞水平上联合分析多模态数据,以鉴定不同的细胞类型。正确的聚类结果对于下游复杂的生物学功能研究至关重要。然而,组合不同的数据源对单细胞多模态数据进行聚类分析仍然是一个统计和计算挑战。在这里,我们开发了一种新颖的多模态深度学习方法scMDC,用于单细胞多组学数据聚类分析。scMDC 是一个端到端的深度模型,它明确表征不同的数据源,并共同学习深度嵌入的潜在特征以进行聚类分析。大量的仿真和真实数据实验表明,scMDC在不同的单细胞多模态数据集上优于现有的单细胞单模态和多模态聚类方法。运行时的线性可扩展性使scMDC成为分析大型多模态数据集的有前途的方法。
2. Github链接
GitHub - xianglin226/scMDC:单细胞多组学深度聚类![]() https://github.com/xianglin226/scMDC
https://github.com/xianglin226/scMDC
二、实验环境
0. 作者要求
Python 3.8.1
Pytorch 1.6.0
Scanpy 1.6.0
SKlearn 0.22.1
Numpy 1.18.1
h5py 2.9.0
- 本研究中scMDC的所有实验都是在Nvidia Tesla P100(16G)GPU上进行的。
- 我们建议在 conda 环境中安装 conda 环境 (conda create -n scMDC)。
- scMDC 对包含 5000 cells的数据集进行聚类大约需要 3分钟。
1. 环境复现
未重新配置环境,继续使用前文深度学习系列文章的环境,实践表明可行:
conda create -n DL python=3.7 conda activate DLpip install torch==1.8.1+cu102 torchvision==0.9.1+cu102 torchaudio==0.8.1 -f https://download.pytorch.org/whl/torch_stable.html
conda install matplotlibconda install scikit-learn实验一
conda install pandasconda install seabornconda install networkxconda install statsmodelspip install pyHSICLasso注:本人的实验环境按照上述顺序安装各种库,若想尝试一起安装(天知道会不会出问题)
实验二(本实验)
pip install scanpy2. 库版本介绍
自己配置的py3.7环境各版本介绍
| 软件包 | 本实验版本 | 目前最新版 |
| matplotlib | 3.5.3 | 3.8.0 |
| numpy | 1.21.6 | 1.26.0 |
| python | 3.7.16 | |
| scikit-learn | 0.22.1 | 1.3.0 |
| torch | 1.8.1+cu102 | 2.0.1 |
| torchaudio | 0.8.1 | 2.0.2 |
| torchvision | 0.9.1+cu102 | 0.15.2 |
实验一
| networkx | 2.6.3 | 3.1 |
| pandas | 1.2.3 | 2.1.1 |
| pyHSICLasso | 1.4.2 | 1.4.2 |
| seaborn | 0.12.2 | 0.13.0 |
| statsmodels | 0.13.5 | 0.14.0 |
实验二
Installing collected packages: stdlib_list, natsort, llvmlite, h5py, session-info, numba, pynndescent, anndata, umap-learn, scanpy
Successfully installed anndata-0.8.0 h5py-3.8.0 llvmlite-0.39.1 natsort-8.4.0 numba-0.56.4 pynndescent-0.5.10 scanpy-1.9.3 session-info-1.0.0 stdlib_list-0.9.0 umap-learn-0.5.43. IDE
建议使用Pycharm
win11 安装 Anaconda(2022.10)+pycharm(2022.3/2023.1.4)+配置虚拟环境_QomolangmaH的博客-CSDN博客https://blog.csdn.net/m0_63834988/article/details/128693741https://blog.csdn.net/m0_63834988/article/details/128693741https://blog.csdn.net/m0_63834988/article/details/128693741![]() https://blog.csdn.net/m0_63834988/article/details/128693741
https://blog.csdn.net/m0_63834988/article/details/128693741
三、实验内容
1. 用法
- 准备 h5 格式的输入数据。(参阅“数据集”文件夹中的自述文件)
- 根据“脚本”文件夹中的运行脚本运行scMDC(如果您处理mRNA+ATAC数据并使用run_scMDC_batch.py进行多批次数据聚类,请注意参数设置)
- 基于训练良好的 scMDC 模型通过run_LRP.py运行 DE 分析(请参阅“脚本”文件夹中的 LRP 运行脚本)
2. 输出
- scMDC输出数据的潜在表示,可用于进一步的下游分析,并通过t-SNE或Umap可视化;
- 多批次 scMDC 输出集成数据集的潜在表示,在其上校正批处理效应。
- LRP输出一个基因等级,指示基因对给定簇的重要性,可用于通路分析。
3. 参数
- --n_clusters:簇数(K);如果此参数设置为 -1,scMDC 将估计 K。
- --cutoff:一个周期的比率,在此之前,模型只训练低级自动编码器。
- --batch_size:批量大小。
- --data_file:数据输入的路径。
- 数据格式:H5。
- 结构:X1(RNA),X2(ADT或ATAC),Y(标签,如果退出),批次(多批次数据聚类的批次指示符)。
- --maxiter:训练的最大时期。默认值:10000。
- --pretrain_epochs:预训练的周期数。默认值:400。
- --gamma:聚类损失系数。默认值:0.1。
- --phi1 和 phi2:预训练和聚类阶段的 KL 损失系数。默认值:CITE-Seq 为 0.001;0.005 表示 SMAGE-Seq*。
- --update_interval:检查性能的间隔。默认值:1。 --tol:停止模型的条件,即更改标签的百分比。
- --tol:0.001。
- --ae_weights:权重文件的路径。
- --save_dir:存储输出的目录。
- --ae_weight_file:存储权重的目录。
- --resolution:用于估计 k 的分辨率参数,默认值:0.2。
- --n_neighbors:估计 K 的n_neighbors参数,默认值:30。
- --embedding_file:如果保存嵌入文件。默认值:否
- --prediction_file:如果保存预测文件。默认值:否
-
--encodeLayer:RNA 的低电平编码器层:默认值:CITE-Seq 的 [256,64,32,16];[256,128,64] 对于 SMAGE-seq。 - --decodeLayer1:ADT 的低级编码器层:默认:CITE-Seq 的 [16,64,256]。[64,128,256] 为 SMAGE-seq。
- --decodeLayer2:高级编码器的层。默认值:[16,20] 表示 CITE-Seq。[64,128,256] 为 SMAGE-seq。
- --sigma1:RNA数据上的噪声。默认值:2.5。
- --sigma2:ADT数据上的噪声。默认值:CITE-Seq 为 1.5;2.5 对于 SMAGE-Seq
- --filter1:如果对基因进行特征选择。默认值:否。
- --filter2:如果在ATAC上进行功能选择。默认值:否。
- --f1:如果进行羽毛选择,则用于聚类的高可变基因(在X1中)的数量。默认值:2000 -
- -f2:如果进行羽毛选择,则来自 ATAC(在 X2 中)的高变量基因数用于聚类。默认值:2000
- *为方便起见,我们将 10X 单细胞多组 ATAC + 基因表达技术表示为 SMAGE-seq。
4. run_scMDC
利用scMultiCluster模型联合利用scRNA-seq和其他模态数据进行细胞类型的无监督聚类,并评估了聚类效果。
设置超参数
读取数据,数据包括两个模态:scRNA-seq读数矩阵X1和另一种基因组数据X2(如ADT/ATAC),以及标签y。
import argparse
parser = argparse.ArgumentParser(description='train',
formatter_class=argparse.ArgumentDefaultsHelpFormatter)
parser.add_argument('--n_clusters', default=12, type=int)
parser.add_argument('--cutoff', default=0.5, type=float,
help='Start to train combined layer after what ratio of epoch')
parser.add_argument('--batch_size', default=256, type=int)
# parser.add_argument('--data_file', default=f'../datasets/SMAGESeq_10X_pbmc_10k_granulocyte_plus.h5')
parser.add_argument('--data_file', default=f'../datasets/output500.h5')
parser.add_argument('--maxiter', default=5000, type=int)
parser.add_argument('--pretrain_epochs', default=200, type=int)
parser.add_argument('--gamma', default=.1, type=float,
help='coefficient of clustering loss')
parser.add_argument('--tau', default=.1, type=float,
help='fuzziness of clustering loss')
parser.add_argument('--phi1', default=0.005, type=float,
help='coefficient of KL loss in pretraining stage')
parser.add_argument('--phi2', default=0.005, type=float,
help='coefficient of KL loss in clustering stage')
parser.add_argument('--update_interval', default=1, type=int)
parser.add_argument('--tol', default=0.001, type=float)
parser.add_argument('--lr', default=1., type=float)
parser.add_argument('--ae_weights', default=None)
parser.add_argument('--save_dir', default='atac_pbmc10k')
parser.add_argument('--ae_weight_file', default='AE_weights_pbmc10k.pth.tar')
parser.add_argument('--resolution', default=0.2, type=float)
parser.add_argument('--n_neighbors', default=30, type=int)
parser.add_argument('--embedding_file', default=True)
parser.add_argument('--prediction_file', default=True)
parser.add_argument('-el', '--encodeLayer', nargs='+', default=[256, 128, 64])
parser.add_argument('-dl1', '--decodeLayer1', nargs='+', default=[64, 128, 256])
parser.add_argument('-dl2', '--decodeLayer2', nargs='+', default=[64, 128, 256])
parser.add_argument('--sigma1', default=2.5, type=float)
parser.add_argument('--sigma2', default=2.5, type=float)
parser.add_argument('--f1', default=1000, type=float, help='Number of mRNA after feature selection')
parser.add_argument('--f2', default=1000, type=float, help='Number of ADT/ATAC after feature selection')
parser.add_argument('--filter1', default=True, help='Do mRNA selection')
parser.add_argument('--filter2', default=True, help='Do ADT/ATAC selection')
parser.add_argument('--run', default=1, type=int)
parser.add_argument('--device', default='cpu')
args = parser.parse_args()
print(args)输出:
Namespace(ae_weight_file='AE_weights_pbmc10k.pth.tar', ae_weights=None, batch_size=256, cutoff=0.5, data_file='../datasets/output500.h5', decodeLayer1=[64, 128, 256], decodeLayer2=[64, 128, 256], device='cpu', embedding_file=True, encodeLayer=[256, 128, 64], f1=1000, f2=1000, filter1=True, filter2=True, gamma=0.1, lr=1.0, maxiter=5000, n_clusters=12, n_neighbors=30, phi1=0.005, phi2=0.005, prediction_file=True, pretrain_epochs=200, resolution=0.2, run=1, save_dir='atac_pbmc10k', sigma1=2.5, sigma2=2.5, tau=0.1, tol=0.001, update_interval=1)对scRNA-seq数据进行预处理
规范化,批量效应校正,log变换等。
data_mat = h5py.File(args.data_file)
x1 = np.array(data_mat['X1'])
x2 = np.array(data_mat['X2'])
y = np.array(data_mat['Y'])
data_mat.close()
# #Gene filter
# if args.filter1:
# importantGenes = geneSelection(x1, n=args.f1, plot=False)
# x1 = x1[:, importantGenes]
# if args.filter2:
# importantGenes = geneSelection(x2, n=args.f2, plot=False)
# x2 = x2[:, importantGenes]
print('------------------------------------')
print(x1.shape)
print(x2.shape)
# preprocessing scRNA-seq read counts matrix
adata1 = sc.AnnData(x1)
adata1.obs['Group'] = y
adata1 = read_dataset(adata1,
transpose=False,
test_split=False,
copy=True)
adata1 = normalize(adata1,
size_factors=True,
normalize_input=True,
filter_min_counts=False,
logtrans_input=True,
nor=0)
adata2 = sc.AnnData(x2)
adata2.obs['Group'] = y
adata2 = read_dataset(adata2,
transpose=False,
test_split=False,
copy=True)
adata2 = normalize(adata2,
size_factors=True,
normalize_input=True,
logtrans_input=True)
# adata2 = clr_normalize_each_cell(adata2)
# adata1.write_h5ad('adata1.h5ad') # 保存adata1为h5ad文件
# adata2.write_h5ad('adata2.h5ad') # 保存adata2为h5ad文件
# print('------------------------------------')
# print(adata1.X.shape)
# print(adata2.X.shape)
input_size1 = adata1.n_vars
input_size2 = adata2.n_vars
print(args)输出:
(11020, 500)
(11020, 500)
Namespace(ae_weight_file='AE_weights_pbmc10k.pth.tar', ae_weights=None, batch_size=256, cutoff=0.5, data_file='../datasets/output500.h5', decodeLayer1=[64, 128, 256], decodeLayer2=[64, 128, 256], device='cpu', embedding_file=True, encodeLayer=[256, 128, 64], f1=1000, f2=1000, filter1=True, filter2=True, gamma=0.1, lr=1.0, maxiter=5000, n_clusters=12, n_neighbors=30, phi1=0.005, phi2=0.005, prediction_file=True, pretrain_epochs=200, resolution=0.2, run=1, save_dir='atac_pbmc10k', sigma1=2.5, sigma2=2.5, tau=0.1, tol=0.001, update_interval=1)构建scMultiCluster模型
- 该模型由一个共享的encoder和两个私有的decoder组成,编码器来连接来自不同模态的数据,并使用两个解码器来单独解码来自每个模态的数据。
- 其中使用 ZINB 损失作为重构损失,因为该损失能很好的表征大量丢失且稀疏的数据,实际应用中已经发现可以有效地拟合scRNA-seq数据并改善表示学习和聚类结果。
- 而使用KL损失可以吸引相似的细胞并分离不同的细胞,帮助得到更好的聚类结果。
- 通过加入Clustering损失能直接针对聚类结果进行优化,实现最优的聚类结果。
encodeLayer = list(map(int, args.encodeLayer))
decodeLayer1 = list(map(int, args.decodeLayer1))
decodeLayer2 = list(map(int, args.decodeLayer2))
model = scMultiCluster(input_dim1=input_size1, input_dim2=input_size2, tau=args.tau,
encodeLayer=encodeLayer, decodeLayer1=decodeLayer1, decodeLayer2=decodeLayer2,
activation='elu', sigma1=args.sigma1, sigma2=args.sigma2, gamma=args.gamma,
cutoff = args.cutoff, phi1=args.phi1, phi2=args.phi2, device=args.device).to(args.device)
print(str(model))
输出:
scMultiCluster(
(encoder): Sequential(
(0): Linear(in_features=1000, out_features=256, bias=True)
(1): BatchNorm1d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ELU(alpha=1.0)
(3): Linear(in_features=256, out_features=128, bias=True)
(4): BatchNorm1d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(5): ELU(alpha=1.0)
(6): Linear(in_features=128, out_features=64, bias=True)
(7): BatchNorm1d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(8): ELU(alpha=1.0)
)
(decoder1): Sequential(
(0): Linear(in_features=64, out_features=128, bias=True)
(1): BatchNorm1d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ELU(alpha=1.0)
(3): Linear(in_features=128, out_features=256, bias=True)
(4): BatchNorm1d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(5): ELU(alpha=1.0)
)
(decoder2): Sequential(
(0): Linear(in_features=64, out_features=128, bias=True)
(1): BatchNorm1d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ELU(alpha=1.0)
(3): Linear(in_features=128, out_features=256, bias=True)
(4): BatchNorm1d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(5): ELU(alpha=1.0)
)
(dec_mean1): Sequential(
(0): Linear(in_features=256, out_features=500, bias=True)
(1): MeanAct()
)
(dec_disp1): Sequential(
(0): Linear(in_features=256, out_features=500, bias=True)
(1): DispAct()
)
(dec_mean2): Sequential(
(0): Linear(in_features=256, out_features=500, bias=True)
(1): MeanAct()
)
(dec_disp2): Sequential(
(0): Linear(in_features=256, out_features=500, bias=True)
(1): DispAct()
)
(dec_pi1): Sequential(
(0): Linear(in_features=256, out_features=500, bias=True)
(1): Sigmoid()
)
(dec_pi2): Sequential(
(0): Linear(in_features=256, out_features=500, bias=True)
(1): Sigmoid()
)
(zinb_loss): ZINBLoss()
)预训练
此时只使用了ZINB损失和KL损失
if not os.path.exists(args.save_dir):
os.makedirs(args.save_dir)
t0 = time()
if args.ae_weights is None:
model.pretrain_autoencoder(X1=adata1.X, X_raw1=adata1.raw.X, sf1=adata1.obs.size_factors,
X2=adata2.X, X_raw2=adata2.raw.X, sf2=adata2.obs.size_factors, batch_size=args.batch_size,
epochs=args.pretrain_epochs, ae_weights=args.ae_weight_file)
else:
if os.path.isfile(args.ae_weights):
print("==> loading checkpoint '{}'".format(args.ae_weights))
checkpoint = torch.load(args.ae_weights)
model.load_state_dict(checkpoint['ae_state_dict'])
else:
print("==> no checkpoint found at '{}'".format(args.ae_weights))
raise ValueError
print('Pretraining time: %d seconds.' % int(time() - t0))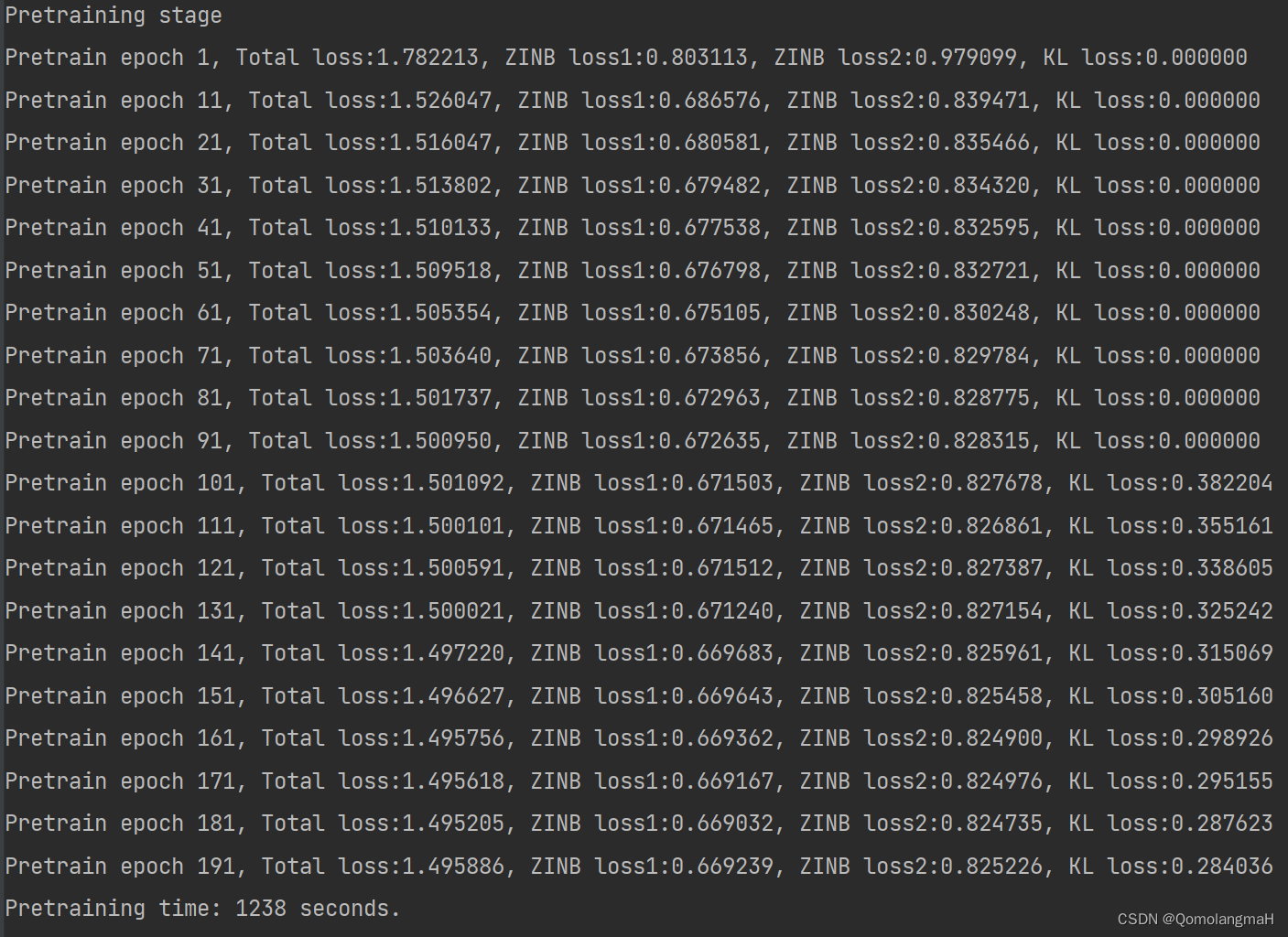
使用KMeans确定聚类数k
#get k
latent = model.encodeBatch(torch.tensor(adata1.X).to(args.device), torch.tensor(adata2.X).to(args.device))
latent = latent.cpu().numpy()
if args.n_clusters == -1:
n_clusters = GetCluster(latent, res=args.resolution, n=args.n_neighbors)
else:
print("n_cluster is defined as " + str(args.n_clusters))
n_clusters = args.n_clustersn_cluster is defined as 12微调模型,训练聚类层
正式的训练过程,此时加入了聚类损失进行模型的训练,从而得到较好的聚类结果
y_pred, _, _, _, _ = model.fit(X1=adata1.X, X_raw1=adata1.raw.X, sf1=adata1.obs.size_factors,
X2=adata2.X, X_raw2=adata2.raw.X, sf2=adata2.obs.size_factors, y=y,
n_clusters=n_clusters, batch_size=args.batch_size, num_epochs=args.maxiter,
update_interval=args.update_interval, tol=args.tol, lr=args.lr,
save_dir=args.save_dir)
print('Total time: %d seconds.' % int(time() - t0))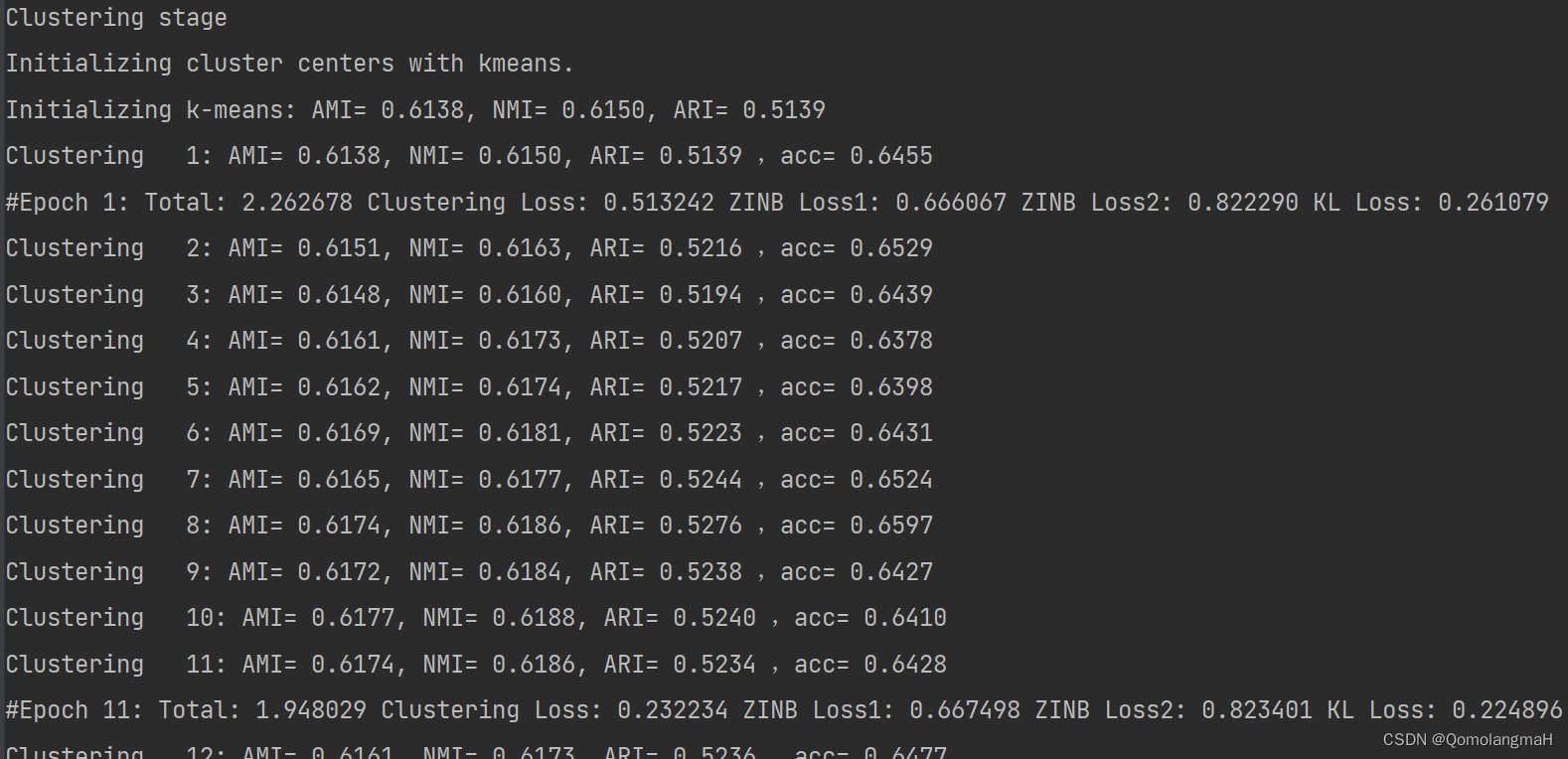
保存预测结果和embedding到文件
if args.prediction_file:
y_pred_ = best_map(y, y_pred) - 1
np.savetxt(args.save_dir + "/" + str(args.run) + "_pred.csv", y_pred_, delimiter=",")
if args.embedding_file:
final_latent = model.encodeBatch(torch.tensor(adata1.X).to(args.device), torch.tensor(adata2.X).to(args.device))
final_latent = final_latent.cpu().numpy()
np.savetxt(args.save_dir + "/" + str(args.run) + "_embedding.csv", final_latent, delimiter=",")

输出预测结果y_pred,计算指标AMI、NMI和ARI
y_pred_ = best_map(y, y_pred)
ami = np.round(metrics.adjusted_mutual_info_score(y, y_pred), 5)
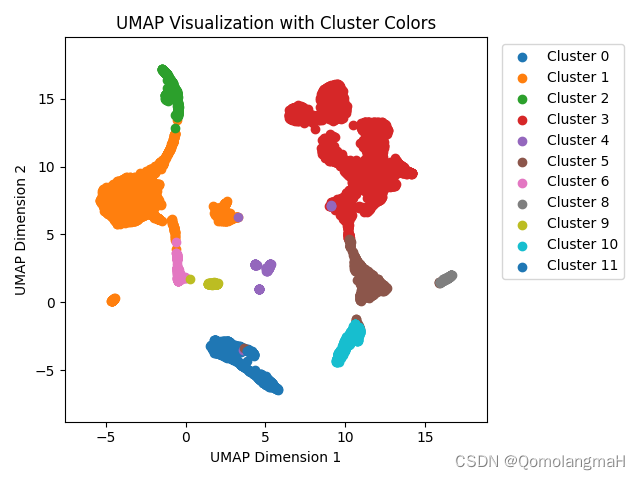
nmi = np.round(metrics.normalized_mutual_info_score(y, y_pred), 5)
ari = np.round(metrics.adjusted_rand_score(y, y_pred), 5)
print('Final: AMI= %.4f, NMI= %.4f, ARI= %.4f' % (ami, nmi, ari))实验结果:
Final: AMI= 0.6565, NMI= 0.6575, ARI= 0.68135. 聚类结果可视化
import umap
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
umap_model = umap.UMAP()
# 读取CSV文件
encoded_data = pd.read_csv('./atac_pbmc10k/1_embedding.csv', header=None)
cluster_labels = pd.read_csv('./atac_pbmc10k/1_pred.csv', header=None)
unique_labels = np.unique(cluster_labels.astype(int))
# 对编码结果进行UMAP转换
umap_data = umap_model.fit_transform(encoded_data)
fig, ax = plt.subplots()
# 使用Matplotlib绘制UMAP转换结果的散点图
for label in unique_labels:
# 提取属于当前簇的数据点的索引
indices = np.where(cluster_labels == label)[0]
# 获取属于当前簇的数据点的UMAP坐标
cluster_points = umap_data[indices, :]
# 使用不同的颜色标记当前簇的数据点
ax.scatter(cluster_points[:, 0], cluster_points[:, 1], label=f'Cluster {label}')
ax.set_xlabel('UMAP Dimension 1')
ax.set_ylabel('UMAP Dimension 2')
ax.set_title('UMAP Visualization with Cluster Colors')
# 显示图例,并将图例放置在右上角的位置
ax.legend(bbox_to_anchor=(1.02, 1), loc='upper left')
x_min, x_max = np.min(umap_data[:, 0]), np.max(umap_data[:, 0])
y_min, y_max = np.min(umap_data[:, 1]), np.max(umap_data[:, 1])
x_margin = (x_max - x_min) * 0.1
y_margin = (y_max - y_min) * 0.1
plt.xlim(x_min - x_margin, x_max + x_margin)
plt.ylim(y_min - y_margin, y_max + y_margin)
# 调整子图布局以缩小UMAP结果的整体尺寸
plt.tight_layout()
# 保存为图片
plt.savefig('umap_visualization.png')
plt.show()
plt.close()可视化结果
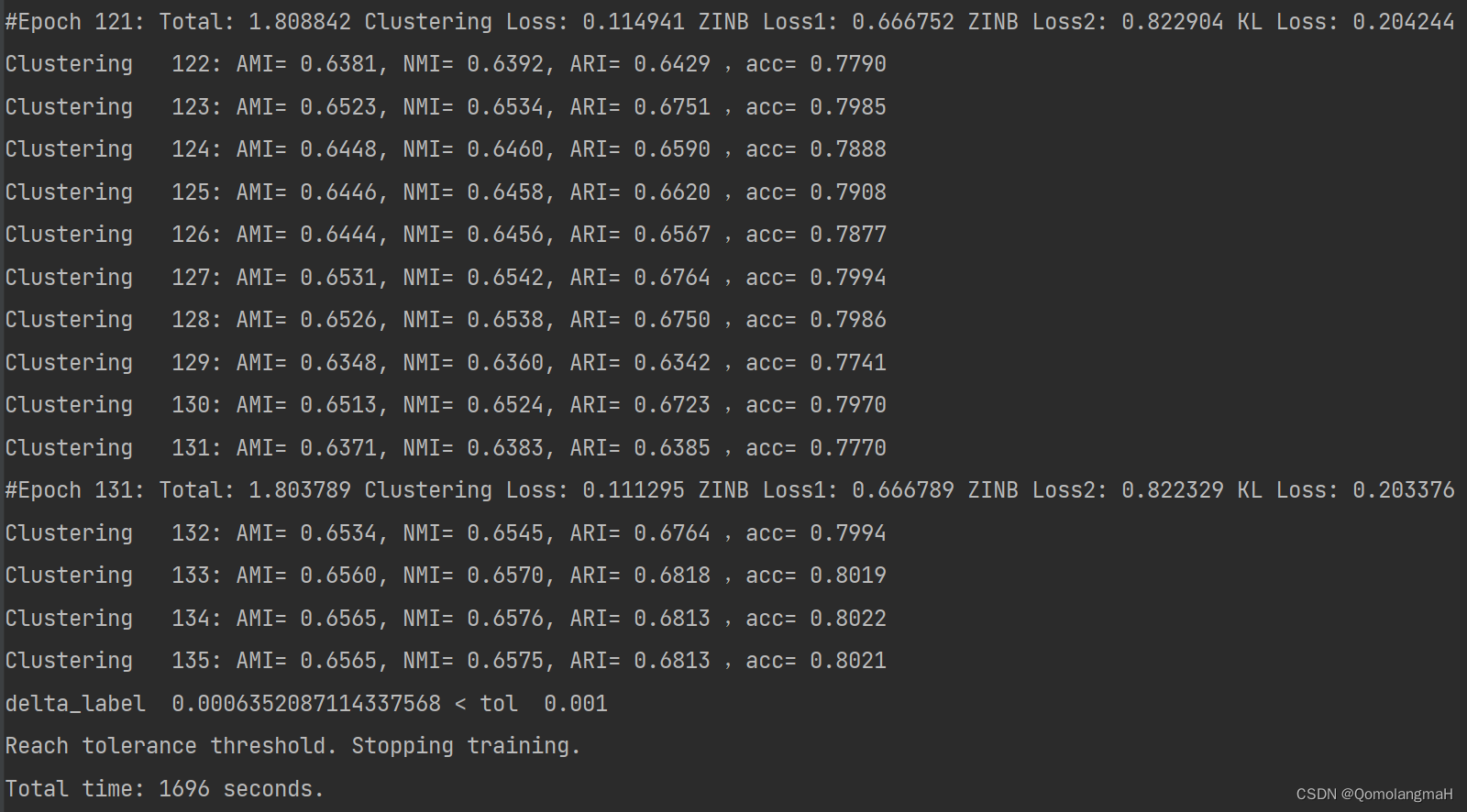
附录:run_scMDC训练过程
### Autoencoder: Successfully preprocessed 500 genes and 11020 cells.
F:\Programming\PycharmProjects\DeepLearning\bio\class2\scMDC-master\scMDC-master\src\run_scMDC.py:106: FutureWarning: X.dtype being converted to np.float32 from float64. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
adata2 = sc.AnnData(x2)
### Autoencoder: Successfully preprocessed 500 genes and 11020 cells.
Namespace(ae_weight_file='AE_weights_pbmc10k.pth.tar', ae_weights=None, batch_size=256, cutoff=0.5, data_file='../datasets/output500.h5', decodeLayer1=[64, 128, 256], decodeLayer2=[64, 128, 256], device='cpu', embedding_file=True, encodeLayer=[256, 128, 64], f1=1000, f2=1000, filter1=True, filter2=True, gamma=0.1, lr=1.0, maxiter=5000, n_clusters=12, n_neighbors=30, phi1=0.005, phi2=0.005, prediction_file=True, pretrain_epochs=200, resolution=0.2, run=1, save_dir='atac_pbmc10k', sigma1=2.5, sigma2=2.5, tau=0.1, tol=0.001, update_interval=1)
scMultiCluster(
(encoder): Sequential(
(0): Linear(in_features=1000, out_features=256, bias=True)
(1): BatchNorm1d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ELU(alpha=1.0)
(3): Linear(in_features=256, out_features=128, bias=True)
(4): BatchNorm1d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(5): ELU(alpha=1.0)
(6): Linear(in_features=128, out_features=64, bias=True)
(7): BatchNorm1d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(8): ELU(alpha=1.0)
)
(decoder1): Sequential(
(0): Linear(in_features=64, out_features=128, bias=True)
(1): BatchNorm1d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ELU(alpha=1.0)
(3): Linear(in_features=128, out_features=256, bias=True)
(4): BatchNorm1d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(5): ELU(alpha=1.0)
)
(decoder2): Sequential(
(0): Linear(in_features=64, out_features=128, bias=True)
(1): BatchNorm1d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ELU(alpha=1.0)
(3): Linear(in_features=128, out_features=256, bias=True)
(4): BatchNorm1d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(5): ELU(alpha=1.0)
)
(dec_mean1): Sequential(
(0): Linear(in_features=256, out_features=500, bias=True)
(1): MeanAct()
)
(dec_disp1): Sequential(
(0): Linear(in_features=256, out_features=500, bias=True)
(1): DispAct()
)
(dec_mean2): Sequential(
(0): Linear(in_features=256, out_features=500, bias=True)
(1): MeanAct()
)
(dec_disp2): Sequential(
(0): Linear(in_features=256, out_features=500, bias=True)
(1): DispAct()
)
(dec_pi1): Sequential(
(0): Linear(in_features=256, out_features=500, bias=True)
(1): Sigmoid()
)
(dec_pi2): Sequential(
(0): Linear(in_features=256, out_features=500, bias=True)
(1): Sigmoid()
)
(zinb_loss): ZINBLoss()
)
Pretraining stage
Pretrain epoch 1, Total loss:1.782213, ZINB loss1:0.803113, ZINB loss2:0.979099, KL loss:0.000000
Pretrain epoch 11, Total loss:1.526047, ZINB loss1:0.686576, ZINB loss2:0.839471, KL loss:0.000000
Pretrain epoch 21, Total loss:1.516047, ZINB loss1:0.680581, ZINB loss2:0.835466, KL loss:0.000000
Pretrain epoch 31, Total loss:1.513802, ZINB loss1:0.679482, ZINB loss2:0.834320, KL loss:0.000000
Pretrain epoch 41, Total loss:1.510133, ZINB loss1:0.677538, ZINB loss2:0.832595, KL loss:0.000000
Pretrain epoch 51, Total loss:1.509518, ZINB loss1:0.676798, ZINB loss2:0.832721, KL loss:0.000000
Pretrain epoch 61, Total loss:1.505354, ZINB loss1:0.675105, ZINB loss2:0.830248, KL loss:0.000000
Pretrain epoch 71, Total loss:1.503640, ZINB loss1:0.673856, ZINB loss2:0.829784, KL loss:0.000000
Pretrain epoch 81, Total loss:1.501737, ZINB loss1:0.672963, ZINB loss2:0.828775, KL loss:0.000000
Pretrain epoch 91, Total loss:1.500950, ZINB loss1:0.672635, ZINB loss2:0.828315, KL loss:0.000000
Pretrain epoch 101, Total loss:1.501092, ZINB loss1:0.671503, ZINB loss2:0.827678, KL loss:0.382204
Pretrain epoch 111, Total loss:1.500101, ZINB loss1:0.671465, ZINB loss2:0.826861, KL loss:0.355161
Pretrain epoch 121, Total loss:1.500591, ZINB loss1:0.671512, ZINB loss2:0.827387, KL loss:0.338605
Pretrain epoch 131, Total loss:1.500021, ZINB loss1:0.671240, ZINB loss2:0.827154, KL loss:0.325242
Pretrain epoch 141, Total loss:1.497220, ZINB loss1:0.669683, ZINB loss2:0.825961, KL loss:0.315069
Pretrain epoch 151, Total loss:1.496627, ZINB loss1:0.669643, ZINB loss2:0.825458, KL loss:0.305160
Pretrain epoch 161, Total loss:1.495756, ZINB loss1:0.669362, ZINB loss2:0.824900, KL loss:0.298926
Pretrain epoch 171, Total loss:1.495618, ZINB loss1:0.669167, ZINB loss2:0.824976, KL loss:0.295155
Pretrain epoch 181, Total loss:1.495205, ZINB loss1:0.669032, ZINB loss2:0.824735, KL loss:0.287623
Pretrain epoch 191, Total loss:1.495886, ZINB loss1:0.669239, ZINB loss2:0.825226, KL loss:0.284036
Pretraining time: 1238 seconds.
n_cluster is defined as 12
Clustering stage
Initializing cluster centers with kmeans.
Initializing k-means: AMI= 0.6138, NMI= 0.6150, ARI= 0.5139
Clustering 1: AMI= 0.6138, NMI= 0.6150, ARI= 0.5139 ,acc= 0.6455
#Epoch 1: Total: 2.262678 Clustering Loss: 0.513242 ZINB Loss1: 0.666067 ZINB Loss2: 0.822290 KL Loss: 0.261079
Clustering 2: AMI= 0.6151, NMI= 0.6163, ARI= 0.5216 ,acc= 0.6529
Clustering 3: AMI= 0.6148, NMI= 0.6160, ARI= 0.5194 ,acc= 0.6439
Clustering 4: AMI= 0.6161, NMI= 0.6173, ARI= 0.5207 ,acc= 0.6378
Clustering 5: AMI= 0.6162, NMI= 0.6174, ARI= 0.5217 ,acc= 0.6398
Clustering 6: AMI= 0.6169, NMI= 0.6181, ARI= 0.5223 ,acc= 0.6431
Clustering 7: AMI= 0.6165, NMI= 0.6177, ARI= 0.5244 ,acc= 0.6524
Clustering 8: AMI= 0.6174, NMI= 0.6186, ARI= 0.5276 ,acc= 0.6597
Clustering 9: AMI= 0.6172, NMI= 0.6184, ARI= 0.5238 ,acc= 0.6427
Clustering 10: AMI= 0.6177, NMI= 0.6188, ARI= 0.5240 ,acc= 0.6410
Clustering 11: AMI= 0.6174, NMI= 0.6186, ARI= 0.5234 ,acc= 0.6428
#Epoch 11: Total: 1.948029 Clustering Loss: 0.232234 ZINB Loss1: 0.667498 ZINB Loss2: 0.823401 KL Loss: 0.224896
Clustering 12: AMI= 0.6161, NMI= 0.6173, ARI= 0.5236 ,acc= 0.6477
Clustering 13: AMI= 0.6163, NMI= 0.6175, ARI= 0.5221 ,acc= 0.6398
Clustering 14: AMI= 0.6162, NMI= 0.6174, ARI= 0.5246 ,acc= 0.6507
Clustering 15: AMI= 0.6162, NMI= 0.6174, ARI= 0.5252 ,acc= 0.6567
Clustering 16: AMI= 0.6160, NMI= 0.6172, ARI= 0.5253 ,acc= 0.6532
Clustering 17: AMI= 0.6171, NMI= 0.6183, ARI= 0.5364 ,acc= 0.6773
Clustering 18: AMI= 0.6163, NMI= 0.6176, ARI= 0.5235 ,acc= 0.6440
Clustering 19: AMI= 0.6179, NMI= 0.6191, ARI= 0.5318 ,acc= 0.6679
Clustering 20: AMI= 0.6161, NMI= 0.6173, ARI= 0.5235 ,acc= 0.6448
Clustering 21: AMI= 0.6174, NMI= 0.6186, ARI= 0.5292 ,acc= 0.6605
#Epoch 21: Total: 1.904330 Clustering Loss: 0.193283 ZINB Loss1: 0.667779 ZINB Loss2: 0.823706 KL Loss: 0.219561
Clustering 22: AMI= 0.6159, NMI= 0.6171, ARI= 0.5233 ,acc= 0.6420
Clustering 23: AMI= 0.6151, NMI= 0.6163, ARI= 0.5231 ,acc= 0.6491
Clustering 24: AMI= 0.6157, NMI= 0.6169, ARI= 0.5229 ,acc= 0.6378
Clustering 25: AMI= 0.6172, NMI= 0.6184, ARI= 0.5334 ,acc= 0.6711
Clustering 26: AMI= 0.6158, NMI= 0.6170, ARI= 0.5265 ,acc= 0.6700
Clustering 27: AMI= 0.6181, NMI= 0.6193, ARI= 0.5386 ,acc= 0.6788
Clustering 28: AMI= 0.6158, NMI= 0.6170, ARI= 0.5239 ,acc= 0.6397
Clustering 29: AMI= 0.6188, NMI= 0.6200, ARI= 0.5383 ,acc= 0.6785
Clustering 30: AMI= 0.6171, NMI= 0.6183, ARI= 0.5255 ,acc= 0.6495
Clustering 31: AMI= 0.6174, NMI= 0.6186, ARI= 0.5264 ,acc= 0.6532
#Epoch 31: Total: 1.881909 Clustering Loss: 0.174528 ZINB Loss1: 0.667321 ZINB Loss2: 0.823386 KL Loss: 0.216674
Clustering 32: AMI= 0.6159, NMI= 0.6171, ARI= 0.5226 ,acc= 0.6438
Clustering 33: AMI= 0.6182, NMI= 0.6194, ARI= 0.5309 ,acc= 0.6648
Clustering 34: AMI= 0.6159, NMI= 0.6171, ARI= 0.5229 ,acc= 0.6410
Clustering 35: AMI= 0.6166, NMI= 0.6178, ARI= 0.5244 ,acc= 0.6443
Clustering 36: AMI= 0.6157, NMI= 0.6169, ARI= 0.5232 ,acc= 0.6404
Clustering 37: AMI= 0.6179, NMI= 0.6190, ARI= 0.5260 ,acc= 0.6488
Clustering 38: AMI= 0.6183, NMI= 0.6195, ARI= 0.5290 ,acc= 0.6600
Clustering 39: AMI= 0.6179, NMI= 0.6191, ARI= 0.5312 ,acc= 0.6658
Clustering 40: AMI= 0.6181, NMI= 0.6193, ARI= 0.5320 ,acc= 0.6681
Clustering 41: AMI= 0.6186, NMI= 0.6198, ARI= 0.5366 ,acc= 0.6754
#Epoch 41: Total: 1.867074 Clustering Loss: 0.161755 ZINB Loss1: 0.667553 ZINB Loss2: 0.823085 KL Loss: 0.214682
Clustering 42: AMI= 0.6179, NMI= 0.6191, ARI= 0.5297 ,acc= 0.6630
Clustering 43: AMI= 0.6173, NMI= 0.6185, ARI= 0.5256 ,acc= 0.6487
Clustering 44: AMI= 0.6150, NMI= 0.6162, ARI= 0.5231 ,acc= 0.6488
Clustering 45: AMI= 0.6175, NMI= 0.6187, ARI= 0.5279 ,acc= 0.6546
Clustering 46: AMI= 0.6186, NMI= 0.6198, ARI= 0.5315 ,acc= 0.6661
Clustering 47: AMI= 0.6180, NMI= 0.6192, ARI= 0.5317 ,acc= 0.6664
Clustering 48: AMI= 0.6171, NMI= 0.6183, ARI= 0.5326 ,acc= 0.6680
Clustering 49: AMI= 0.6186, NMI= 0.6198, ARI= 0.5321 ,acc= 0.6668
Clustering 50: AMI= 0.6171, NMI= 0.6183, ARI= 0.5286 ,acc= 0.6609
Clustering 51: AMI= 0.6186, NMI= 0.6198, ARI= 0.5317 ,acc= 0.6655
#Epoch 51: Total: 1.854746 Clustering Loss: 0.151936 ZINB Loss1: 0.667286 ZINB Loss2: 0.822858 KL Loss: 0.212667
Clustering 52: AMI= 0.6165, NMI= 0.6177, ARI= 0.5271 ,acc= 0.6545
Clustering 53: AMI= 0.6151, NMI= 0.6163, ARI= 0.5232 ,acc= 0.6374
Clustering 54: AMI= 0.6173, NMI= 0.6185, ARI= 0.5273 ,acc= 0.6571
Clustering 55: AMI= 0.6160, NMI= 0.6172, ARI= 0.5227 ,acc= 0.6375
Clustering 56: AMI= 0.6151, NMI= 0.6163, ARI= 0.5230 ,acc= 0.6504
Clustering 57: AMI= 0.6190, NMI= 0.6202, ARI= 0.5405 ,acc= 0.6825
Clustering 58: AMI= 0.6180, NMI= 0.6192, ARI= 0.5296 ,acc= 0.6625
Clustering 59: AMI= 0.6159, NMI= 0.6171, ARI= 0.5231 ,acc= 0.6391
Clustering 60: AMI= 0.6174, NMI= 0.6186, ARI= 0.5297 ,acc= 0.6619
Clustering 61: AMI= 0.6176, NMI= 0.6188, ARI= 0.5277 ,acc= 0.6547
#Epoch 61: Total: 1.844312 Clustering Loss: 0.143666 ZINB Loss1: 0.667285 ZINB Loss2: 0.822651 KL Loss: 0.210710
Clustering 62: AMI= 0.6178, NMI= 0.6190, ARI= 0.5306 ,acc= 0.6644
Clustering 63: AMI= 0.6197, NMI= 0.6209, ARI= 0.5392 ,acc= 0.6800
Clustering 64: AMI= 0.6153, NMI= 0.6165, ARI= 0.5229 ,acc= 0.6436
Clustering 65: AMI= 0.6148, NMI= 0.6160, ARI= 0.5224 ,acc= 0.6383
Clustering 66: AMI= 0.6150, NMI= 0.6162, ARI= 0.5228 ,acc= 0.6380
Clustering 67: AMI= 0.6173, NMI= 0.6186, ARI= 0.5262 ,acc= 0.6549
Clustering 68: AMI= 0.6163, NMI= 0.6175, ARI= 0.5235 ,acc= 0.6450
Clustering 69: AMI= 0.6155, NMI= 0.6167, ARI= 0.5227 ,acc= 0.6414
Clustering 70: AMI= 0.6182, NMI= 0.6194, ARI= 0.5315 ,acc= 0.6672
Clustering 71: AMI= 0.6177, NMI= 0.6189, ARI= 0.5292 ,acc= 0.6623
#Epoch 71: Total: 1.837503 Clustering Loss: 0.137466 ZINB Loss1: 0.667154 ZINB Loss2: 0.823006 KL Loss: 0.209877
Clustering 72: AMI= 0.6193, NMI= 0.6205, ARI= 0.5354 ,acc= 0.6741
Clustering 73: AMI= 0.6178, NMI= 0.6190, ARI= 0.5271 ,acc= 0.6565
Clustering 74: AMI= 0.6164, NMI= 0.6176, ARI= 0.5283 ,acc= 0.6595
Clustering 75: AMI= 0.6205, NMI= 0.6217, ARI= 0.5467 ,acc= 0.6907
Clustering 76: AMI= 0.6184, NMI= 0.6196, ARI= 0.5365 ,acc= 0.6762
Clustering 77: AMI= 0.6162, NMI= 0.6175, ARI= 0.5233 ,acc= 0.6411
Clustering 78: AMI= 0.6165, NMI= 0.6177, ARI= 0.5250 ,acc= 0.6468
Clustering 79: AMI= 0.6182, NMI= 0.6194, ARI= 0.5325 ,acc= 0.6694
Clustering 80: AMI= 0.6148, NMI= 0.6160, ARI= 0.5219 ,acc= 0.6413
Clustering 81: AMI= 0.6155, NMI= 0.6167, ARI= 0.5226 ,acc= 0.6381
#Epoch 81: Total: 1.829174 Clustering Loss: 0.131217 ZINB Loss1: 0.666992 ZINB Loss2: 0.822744 KL Loss: 0.208221
Clustering 82: AMI= 0.6169, NMI= 0.6181, ARI= 0.5292 ,acc= 0.6621
Clustering 83: AMI= 0.6161, NMI= 0.6173, ARI= 0.5231 ,acc= 0.6398
Clustering 84: AMI= 0.6174, NMI= 0.6186, ARI= 0.5304 ,acc= 0.6645
Clustering 85: AMI= 0.6143, NMI= 0.6155, ARI= 0.5223 ,acc= 0.6382
Clustering 86: AMI= 0.6182, NMI= 0.6194, ARI= 0.5368 ,acc= 0.6767
Clustering 87: AMI= 0.6189, NMI= 0.6201, ARI= 0.5410 ,acc= 0.6833
Clustering 88: AMI= 0.6162, NMI= 0.6174, ARI= 0.5242 ,acc= 0.6465
Clustering 89: AMI= 0.6157, NMI= 0.6169, ARI= 0.5253 ,acc= 0.6517
Clustering 90: AMI= 0.6170, NMI= 0.6182, ARI= 0.5278 ,acc= 0.6595
Clustering 91: AMI= 0.6166, NMI= 0.6178, ARI= 0.5306 ,acc= 0.6647
#Epoch 91: Total: 1.822761 Clustering Loss: 0.126242 ZINB Loss1: 0.666825 ZINB Loss2: 0.822552 KL Loss: 0.207142
Clustering 92: AMI= 0.6182, NMI= 0.6194, ARI= 0.5399 ,acc= 0.6824
Clustering 93: AMI= 0.6136, NMI= 0.6148, ARI= 0.5246 ,acc= 0.6499
Clustering 94: AMI= 0.6169, NMI= 0.6182, ARI= 0.5400 ,acc= 0.6822
Clustering 95: AMI= 0.6180, NMI= 0.6192, ARI= 0.5478 ,acc= 0.6946
Clustering 96: AMI= 0.6080, NMI= 0.6093, ARI= 0.5229 ,acc= 0.6466
Clustering 97: AMI= 0.6151, NMI= 0.6163, ARI= 0.5506 ,acc= 0.6985
Clustering 98: AMI= 0.6151, NMI= 0.6163, ARI= 0.5461 ,acc= 0.6935
Clustering 99: AMI= 0.6134, NMI= 0.6146, ARI= 0.5449 ,acc= 0.6916
Clustering 100: AMI= 0.6119, NMI= 0.6131, ARI= 0.5476 ,acc= 0.6966
Clustering 101: AMI= 0.6103, NMI= 0.6115, ARI= 0.5355 ,acc= 0.6808
#Epoch 101: Total: 1.817155 Clustering Loss: 0.122029 ZINB Loss1: 0.666804 ZINB Loss2: 0.822372 KL Loss: 0.205949
Clustering 102: AMI= 0.6162, NMI= 0.6174, ARI= 0.5599 ,acc= 0.7123
Clustering 103: AMI= 0.6121, NMI= 0.6134, ARI= 0.5448 ,acc= 0.6955
Clustering 104: AMI= 0.6194, NMI= 0.6207, ARI= 0.5718 ,acc= 0.7265
Clustering 105: AMI= 0.6128, NMI= 0.6140, ARI= 0.5524 ,acc= 0.7064
Clustering 106: AMI= 0.6159, NMI= 0.6171, ARI= 0.5718 ,acc= 0.7266
Clustering 107: AMI= 0.6322, NMI= 0.6334, ARI= 0.6308 ,acc= 0.7708
Clustering 108: AMI= 0.6300, NMI= 0.6312, ARI= 0.6223 ,acc= 0.7665
Clustering 109: AMI= 0.6290, NMI= 0.6302, ARI= 0.6208 ,acc= 0.7643
Clustering 110: AMI= 0.6233, NMI= 0.6245, ARI= 0.6049 ,acc= 0.7538
Clustering 111: AMI= 0.6494, NMI= 0.6506, ARI= 0.6700 ,acc= 0.7955
#Epoch 111: Total: 1.812602 Clustering Loss: 0.118209 ZINB Loss1: 0.666840 ZINB Loss2: 0.822511 KL Loss: 0.205043
Clustering 112: AMI= 0.6432, NMI= 0.6444, ARI= 0.6577 ,acc= 0.7883
Clustering 113: AMI= 0.6419, NMI= 0.6431, ARI= 0.6555 ,acc= 0.7860
Clustering 114: AMI= 0.6212, NMI= 0.6224, ARI= 0.5924 ,acc= 0.7449
Clustering 115: AMI= 0.6342, NMI= 0.6354, ARI= 0.6385 ,acc= 0.7747
Clustering 116: AMI= 0.6132, NMI= 0.6144, ARI= 0.5697 ,acc= 0.7258
Clustering 117: AMI= 0.6266, NMI= 0.6278, ARI= 0.6163 ,acc= 0.7622
Clustering 118: AMI= 0.6493, NMI= 0.6505, ARI= 0.6698 ,acc= 0.7952
Clustering 119: AMI= 0.6267, NMI= 0.6279, ARI= 0.6201 ,acc= 0.7635
Clustering 120: AMI= 0.6365, NMI= 0.6378, ARI= 0.6473 ,acc= 0.7811
Clustering 121: AMI= 0.6457, NMI= 0.6469, ARI= 0.6624 ,acc= 0.7910
#Epoch 121: Total: 1.808842 Clustering Loss: 0.114941 ZINB Loss1: 0.666752 ZINB Loss2: 0.822904 KL Loss: 0.204244
Clustering 122: AMI= 0.6381, NMI= 0.6392, ARI= 0.6429 ,acc= 0.7790
Clustering 123: AMI= 0.6523, NMI= 0.6534, ARI= 0.6751 ,acc= 0.7985
Clustering 124: AMI= 0.6448, NMI= 0.6460, ARI= 0.6590 ,acc= 0.7888
Clustering 125: AMI= 0.6446, NMI= 0.6458, ARI= 0.6620 ,acc= 0.7908
Clustering 126: AMI= 0.6444, NMI= 0.6456, ARI= 0.6567 ,acc= 0.7877
Clustering 127: AMI= 0.6531, NMI= 0.6542, ARI= 0.6764 ,acc= 0.7994
Clustering 128: AMI= 0.6526, NMI= 0.6538, ARI= 0.6750 ,acc= 0.7986
Clustering 129: AMI= 0.6348, NMI= 0.6360, ARI= 0.6342 ,acc= 0.7741
Clustering 130: AMI= 0.6513, NMI= 0.6524, ARI= 0.6723 ,acc= 0.7970
Clustering 131: AMI= 0.6371, NMI= 0.6383, ARI= 0.6385 ,acc= 0.7770
#Epoch 131: Total: 1.803789 Clustering Loss: 0.111295 ZINB Loss1: 0.666789 ZINB Loss2: 0.822329 KL Loss: 0.203376
Clustering 132: AMI= 0.6534, NMI= 0.6545, ARI= 0.6764 ,acc= 0.7994
Clustering 133: AMI= 0.6560, NMI= 0.6570, ARI= 0.6818 ,acc= 0.8019
Clustering 134: AMI= 0.6565, NMI= 0.6576, ARI= 0.6813 ,acc= 0.8022
Clustering 135: AMI= 0.6565, NMI= 0.6575, ARI= 0.6813 ,acc= 0.8021
delta_label 0.0006352087114337568 < tol 0.001
Reach tolerance threshold. Stopping training.
Total time: 1696 seconds.
Final: AMI= 0.6565, NMI= 0.6575, ARI= 0.6813





![[python 刷题] 19 Remove Nth Node From End of List](https://img-blog.csdnimg.cn/07b6189a539e4dd2bf62f3e5a7a7e8a8.jpeg#pic_center)