目录
01 实验题目
02 我的IDL代码
03 实验给定IDL代码
04 Python代码实现
4.1 我的Python代码
4.2 实验给定Python代码
01 实验题目
计算/coarse_data/chapter_2/NO2/目录下所有OMI-NO2产品数据集ColumnAmountNO2TropCloudScreened的月均值、季均值、年均值,并以Geotiff形式输出,输出单位为mol/km2。要求程序一次完成所有均值的计算与输出,并尝试不设置起始年和年份数,实现年份的自动统计。
注:月均值、季均值不分年份;春季为3-5月,夏季为6-8月,秋季为9-11月,冬季为12-2月。
由于我的做法和给定实验的代码思路不完全一致,思路上稍微有些差别,但是核心都是一样的,最终处理的结果几乎一致。
我的核心思路:
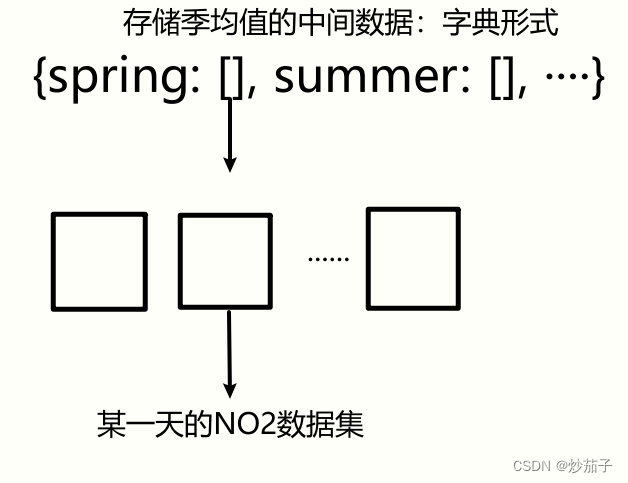
创建三个字典(在IDL中为哈希表, 实际类似于Python的字典),分别用于存储月均值、季均值、年均值的中间数据,类似下方:
通过HDF5文件名知晓该文件的日期进而获取其属于年?季?月等信息,然后分配到不同的[]中,[]在图示中不是数组而是列表(在IDL中为链表,实际类似于Python的列表), 由于我们事先不知道春季有多少文件,所以定义列表更为合适(只需要add添加元素<此处为数组>即可)。列表中可以存储多个数组,一个数组即为某一天的NO2数据集。
如何求取均值呢?
好问题,我们先将例如上方的spring的列表转化为数组(其是三维数组,分别表示数据集的行列数和数据集的个数),接着基于数据集个数这一维度使用mean函数进行求取均值即可.
实验给定代码:
其是通过计算每一像元的累积和以及该像元的实际有效天数或者次数进行均值的求取,对于像元的有效次数的求取在我的代码中通过mean函数的/nan参数解决,自动跳过无效值(所以需要事先将所有无效值替换为NAN),其余基本一致。
02 我的IDL代码
function extract_start_end, files_path, files_amount=files_amount
; 该函数用于从存储多个路径数组中获取年份信息并返回起始-终止年份
years = list()
for i = 0, files_amount - 1 do begin
year = fix(strmid(file_basename(files_path[i]), 19, 4))
if where(years eq year) EQ -1 then years.add, year
endfor
years = years.toarray()
return, [min(years), max(years)]
end
function read_h5, h5_path, group_path=group_path, ds_name=ds_name
; 该函数用于读取HDF5文件的数据集
; 如果关键字参数没有传入, 设置默认
if ~keyword_set(group_path) then group_path = 'HDFEOS/GRIDS/ColumnAmountNO2/Data Fields'
if ~keyword_set(ds_name) then ds_name = 'ColumnAmountNO2TropCloudScreened'
file_id = h5f_open(h5_path) ; 默认可读模式打开, 返回文件ID(指针)
group_id = h5g_open(file_id, group_path) ; 获取组ID
ds_id = h5d_open(group_id, ds_name)
ds = h5d_read(ds_id) ; 获取数据集
; 关闭以释放资源
h5d_close, ds_id
h5g_close, group_id
h5f_close, file_id
return, ds
end
pro OMI_NO2_mean
; 此程序用于计算OMI-NO2产品下ColumnAmountNO2TropCloudScreened数据集的$
; 月均值、 季均值、 年均值, 并以Geotiff形式输出, 输出单位为mol/km2
start_time = systime(1) ; 记录时间
; 准备
in_dir = 'D:\Objects\JuniorFallTerm\IDLProgram\Experiments\ExperimentalData\chapter_2'
out_dir = 'D:\Objects\JuniorFallTerm\IDLProgram\Experiments\ExperimentalData\chapter_2\Output'
if ~file_test(out_dir, /directory) then file_mkdir, out_dir ; 是否存在该文件夹, 不存在创建
; 基本信息
ds_dir = 'HDFEOS/GRIDS/ColumnAmountNO2/Data Fields'
ds_name = 'ColumnAmountNO2TropCloudScreened'
rows = 720
cols = 1440
season_info = hash(3, 'spring', 4, 'spring', 5, 'spring',$
6, 'summer', 7, 'summer', 8, 'summer', 9, 'autumn', 10, 'autumn',$
11, 'autumn', 12, 'winter', 1, 'winter', 2, 'winter')
; 创建哈希表进行月份与季节(key)的对应
; 获取起始-终止年份
files_path = file_search(in_dir, 'OMI-Aura_L3*.he5', count=files_amount) ; 查询所有满足条件的文件路径
start_end_year = extract_start_end(files_path, files_amount=files_amount)
years_amount = start_end_year[-1] - start_end_year[0] + 1 ; 获取年份数目
; 创建存储池
year_box = hash() ; 哈希表, 类似python-字典
month_box = hash()
season_box = hash()
for i = start_end_year[0], start_end_year[-1] do year_box[i] = list() ; list类似python-列表
for i = 1, 12 do month_box[i] = list()
foreach i, ['spring', 'summer', 'autumn', 'winter'] do season_box[i] = list()
; 循环每一个HDF5文件
for file_i = 0, files_amount - 1 do begin
; 获取当前循环的HDF5文件路径及基本信息
df_path = files_path[file_i]
df_year = fix(strmid(file_basename(df_path), 19, 4))
df_month = fix(strmid(file_basename(df_path), 24, 2))
df_season = season_info[df_month]
; 读取对流层NO2的垂直柱含量及基本处理
ds = read_h5(df_path)
ds[where(ds lt 0, /null)] = !values.F_NAN
; 单位换算molec/cm^2 ==> mol/km^2, 1mol = 6.022 * 10 ^ 23(即NA), 1km^2 = 10 ^ 10 cm^2
ds = (ds * 10.0 ^ 10) / !const.NA ; !const.NA = 6.022 * 10 ^ 23
; 上方为北极(由于此极轨卫星从南极拍摄, 故影像第一行为南极位置的第一行, 需南北颠倒)
ds = rotate(ds, 7) ; 7: x ==> x, y ==> -y
; 如果自己写不用函数或可
; ds = ds[*, rows - indgen(rows, start=1)]
; 加和
year_box[df_year].add, ds
month_box[df_month].add, ds
season_box[df_season].add, ds
endfor
; 投影信息
geo_info={$
MODELPIXELSCALETAG:[0.25,0.25,0.0],$ ; 经度分辨率, 维度分辨率, 高程分辨率(Z轴) ==> 不知前面是否反了
MODELTIEPOINTTAG:[0.0,0.0,0.0,-180.0,90.0,0.0],$ ; 第0列第0行第0高的像元点的经纬度高程分别为 -180, 90, 0
GTMODELTYPEGEOKEY:2,$ ; 设置为地理坐标系
GTRASTERTYPEGEOKEY:1,$ ; 像素的表示类型, 北上图像(North-Up)
GEOGRAPHICTYPEGEOKEY:4326,$ ; 地理坐标系为WGS84
GEOGCITATIONGEOKEY:'GCS_WGS_1984',$
GEOGANGULARUNITSGEOKEY:9102,$ ; 单位为度
GEOGSEMIMAJORAXISGEOKEY:6378137.0,$ ; 主半轴长度为6378137.0m
GEOGINVFLATTENINGGEOKEY:298.25722} ; 反扁平率为298.25722
; 计算均值(求取均值警告存在Floating illegal operand是由于某一像元位置任意时间上均为NAN导致, 不影响输出结果)
for i = start_end_year[0], start_end_year[-1] do begin
year_box[i] = mean(year_box[i].toarray(), dimensio=1, /nan) ; dimension=1表示第一个维度(索引从1开始)
year_path = out_dir + '\year_mean_' + strcompress(string(i), /remove_all) + '.tiff'
write_tiff, year_path, year_box[i], geotiff=geo_info, /float
endfor
for i = 1, 12 do begin
month_box[i] = mean(month_box[i].toarray(), dimension=1, /nan)
month_path = out_dir + '\month_mean_' + strcompress(string(i), /remove_all) + '.tiff'
write_tiff, month_path, month_box[i], geotiff=geo_info, /float
endfor
foreach i, ['spring', 'summer', 'autumn', 'winter'] do begin
season_box[i] = mean(season_box[i].toarray(), dimension=1, /nan)
season_path = out_dir + '\season_mean_' + i + '.tiff'
write_tiff, season_path, season_box[i], geotiff=geo_info, /float
endforeach
end_time = systime(1)
print, end_time - start_time, format="均值处理完成, 用时: %6.2f s"
end03 实验给定IDL代码
function h5_data_get,file_name,dataset_name
file_id=h5f_open(file_name)
dataset_id=h5d_open(file_id,dataset_name)
data=h5d_read(dataset_id)
h5d_close,dataset_id
h5f_close,file_id
return,data
data=!null
end
pro omi_no2_average_calculating
;输入输出路径设置
start_time=systime(1)
in_path='O:/coarse_data/chapter_2/NO2/'
out_path='O:/coarse_data/chapter_2/NO2/average/'
dir_test=file_test(out_path,/directory)
if dir_test eq 0 then begin
file_mkdir,out_path
endif
filelist=file_search(in_path,'*NO2*.he5')
file_n=n_elements(filelist)
group_name='/HDFEOS/GRIDS/ColumnAmountNO2/Data Fields/'
target_dataset='ColumnAmountNO2TropCloudScreened'
dataset_name=group_name+target_dataset
;print,dataset_name
;月份存储数组初始化
data_total_month=fltarr(1440,720,12)
data_valid_month=fltarr(1440,720,12)
;季节存储数组初始化
data_total_season=fltarr(1440,720,4)
data_valid_season=fltarr(1440,720,4)
;处理年份设置、年份存储数组初始化
year_start=2017
year_n=2
data_total_year=fltarr(1440,720,year_n)
data_valid_year=fltarr(1440,720,year_n)
for file_i=0,file_n-1 do begin
data_temp=h5_data_get(filelist[file_i],dataset_name)
data_temp=((data_temp gt 0.0)*data_temp/!const.NA)*(10.0^10.0);转mol/km2
data_temp=rotate(data_temp,7)
layer_i=fix(strmid(file_basename(filelist[file_i]),24,2))-1
data_total_month[*,*,layer_i]+= data_temp
data_valid_month[*,*,layer_i]+= (data_temp gt 0.0)
if (layer_i ge 2) and (layer_i le 4) then begin
data_total_season[*,*,0]+=data_temp
data_valid_season[*,*,0]+=(data_temp gt 0.0)
endif
if (layer_i ge 5) and (layer_i le 7) then begin
data_total_season[*,*,1]+=data_temp
data_valid_season[*,*,1]+=(data_temp gt 0.0)
endif
if (layer_i ge 8) and (layer_i le 10) then begin
data_total_season[*,*,2]+=data_temp
data_valid_season[*,*,2]+=(data_temp gt 0.0)
endif
if (layer_i ge 11) or (layer_i le 1) then begin
data_total_season[*,*,3]+=data_temp
data_valid_season[*,*,3]+=(data_temp gt 0.0)
endif
year_i=fix(strmid(file_basename(filelist[file_i]),19,4))-year_start
data_total_year[*,*,year_i]+=data_temp
data_valid_year[*,*,year_i]+=data_temp
endfor
data_valid_month=(data_valid_month gt 0.0)*data_valid_month+(data_valid_month eq 0.0)*(1.0)
data_avr_month=data_total_month/data_valid_month
data_valid_season=(data_valid_season gt 0.0)*data_valid_season+(data_valid_season eq 0.0)*(1.0)
data_avr_season=data_total_season/data_valid_season
data_valid_year=(data_valid_year gt 0.0)*data_valid_year+(data_valid_year eq 0.0)*(1.0)
data_avr_year=data_total_year/data_valid_year
month_out=['01','02','03','04','05','06','07','08','09','10','11','12']
season_out=['spring','summer','autumn','winter']
year_out=['2017','2018']
geo_info={$
MODELPIXELSCALETAG:[0.25,0.25,0.0],$
MODELTIEPOINTTAG:[0.0,0.0,0.0,-180.0,90.0,0.0],$
GTMODELTYPEGEOKEY:2,$
GTRASTERTYPEGEOKEY:1,$
GEOGRAPHICTYPEGEOKEY:4326,$
GEOGCITATIONGEOKEY:'GCS_WGS_1984',$
GEOGANGULARUNITSGEOKEY:9102,$
GEOGSEMIMAJORAXISGEOKEY:6378137.0,$
GEOGINVFLATTENINGGEOKEY:298.25722}
for month_i=0,11 do begin
out_name=out_path+'month_avr_'+month_out[month_i]+'.tiff'
write_tiff,out_name,data_avr_month[*,*,month_i],/float,geotiff=geo_info
endfor
for season_i=0,3 do begin
out_name=out_path+'month_avr_'+season_out[season_i]+'.tiff'
write_tiff,out_name,data_avr_season[*,*,season_i],/float,geotiff=geo_info
endfor
for year_i=0,1 do begin
out_name=out_path+'year_avr_'+year_out[year_i]+'.tiff'
write_tiff,out_name,data_avr_year[*,*,year_i],/float,geotiff=geo_info
endfor
end_time=systime(1)
print,'Processing is end, the totol time consumption is:'+strcompress(string(end_time-start_time))+' s.'
end
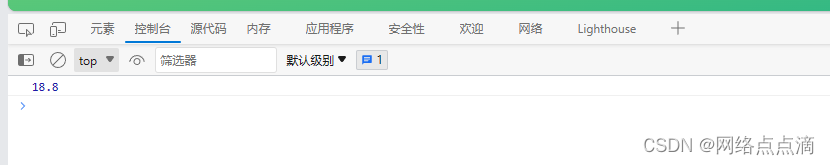
二者对比(仅对比2017,其他结果查验均类似如此):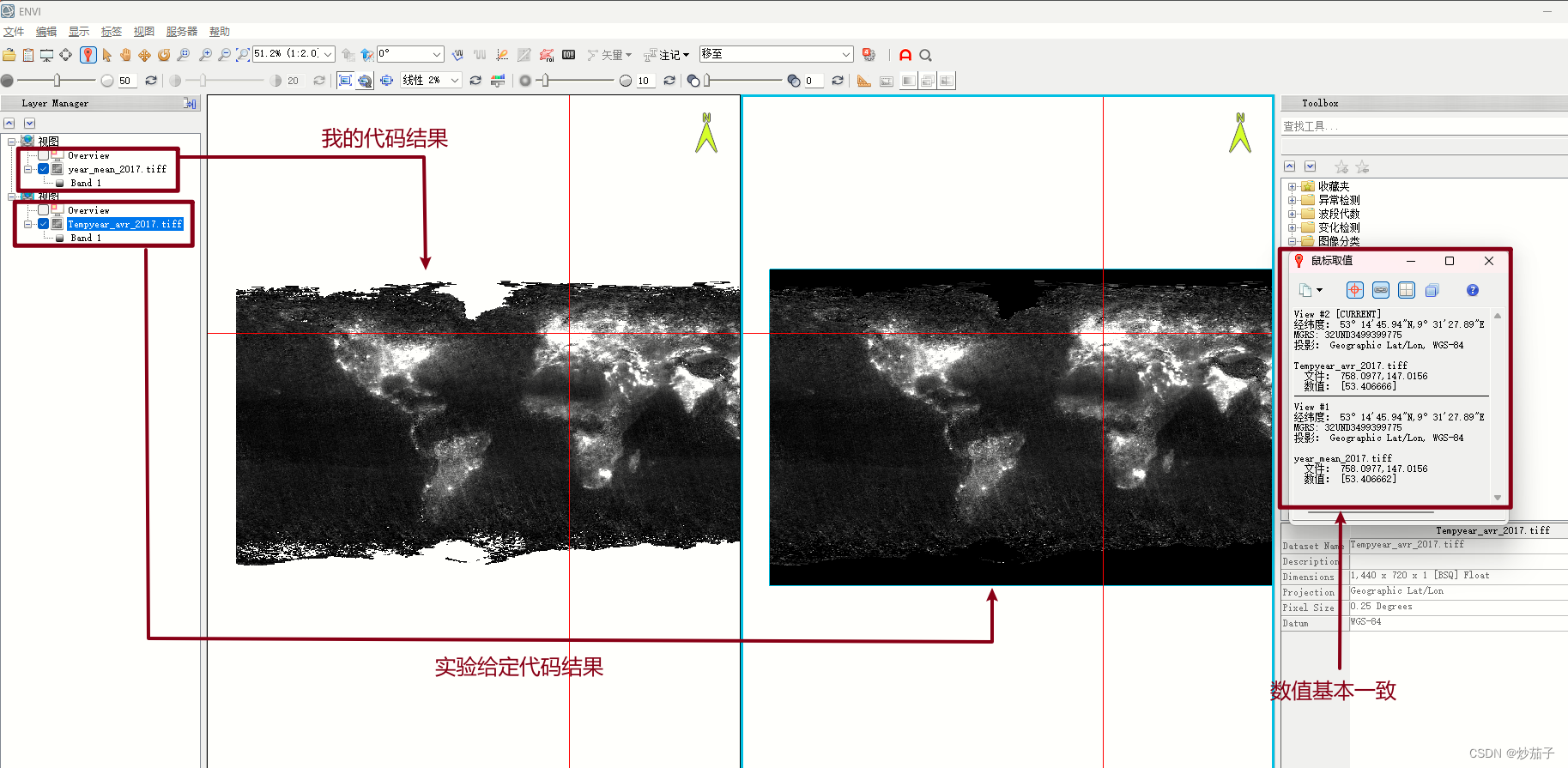
04 Python代码实现
4.1 我的Python代码
import os
import glob
import numpy as np
from scipy import constants
import h5py
from osgeo import gdal
def write_tiff(out_path, data, geotrans, rows=720, cols=1440, bands=1):
driver = gdal.GetDriverByName('GTiff')
df = driver.Create(out_path, cols, rows, bands, gdal.GDT_Float32)
df.SetGeoTransform(geotrans)
df.SetProjection('WGS84')
df.GetRasterBand(1).WriteArray(data)
df.FlushCache()
df = None
# 准备工作
in_dir = r'D:\Objects\JuniorFallTerm\IDLProgram\Experiments\ExperimentalData\chapter_2'
geotrans = (-180, 0.25, 0, 90, 0, -0.25) # 仿射变换参数: (左上角经度, 经度分辨率, 旋转角度, 左上角纬度, 旋转角度, 纬度分辨率)
# 读取数据
file_paths = glob.glob(os.path.join(in_dir, r'**\OMI-Aura_L3*.he5'), recursive=True) # recursive=True表示递归查找,**表示任意个子目录
years = set([int(os.path.basename(file)[19:23]) for file in file_paths]) # 从文件名中提取年份
year_box = {year: [] for year in years} # 用于存放每年的数据
season_box = {season: [] for season in ['winter', 'spring', 'summer', 'autumn']} # 用于存放每季的数据
month_box = {month: [] for month in range(1, 13)} # 用于存放每月的数据
season_info = {1: 'winter', 2: 'winter', 3: 'spring', 4: 'spring', 5: 'spring', 6: 'summer', 7: 'summer', 8: 'summer', 9: 'autumn', 10: 'autumn', 11: 'autumn', 12: 'winter'}
for path in file_paths:
year = int(os.path.basename(path)[19:23])
month = int(os.path.basename(path)[24:26])
season = season_info[month]
with h5py.File(path, 'r') as f:
data = f['HDFEOS/GRIDS/ColumnAmountNO2/Data Fields/ColumnAmountNO2TropCloudScreened'][:]
# 缺失值处理
data[data < 0] = np.nan
# 阿伏伽德罗常数也可以使用scipy.constants中的value
data = (data * 10 ** 10) / constants.Avogadro
# 北极在上
data = np.flipud(data)
year_box[year].append(data)
season_box[season].append(data)
month_box[month].append(data)
# 写入文件
out_dir = r'D:\Objects\JuniorFallTerm\IDLProgram\Experiments\ExperimentalData\chapter_2\PyOutput'
if not os.path.exists(out_dir):
os.makedirs(out_dir)
for year, data in year_box.items():
out_path = os.path.join(out_dir, f'OMI_NO2_{year}_mean.tif')
write_tiff(out_path, np.nanmean(np.array(data), axis=0), geotrans) # RuntimeWarning: Mean of empty slice是因为某一像元所有位置均为NAN
for season, data in season_box.items():
out_path = os.path.join(out_dir, f'OMI_NO2_{season}_mean.tif')
write_tiff(out_path, np.nanmean(np.array(data), axis=0), geotrans)
for month, data in month_box.items():
out_path = os.path.join(out_dir, f'OMI_NO2_{month}_mean.tif')
write_tiff(out_path, np.nanmean(np.array(data), axis=0), geotrans)
4.2 实验给定Python代码
import os
import numpy as np
import h5py as h5
from scipy import constants as ct
from osgeo import gdal
from osgeo import osr
def main():
input_directory = 'O:/coarse_data/chapter_2/NO2/'
output_directory = 'O:/coarse_data/chapter_2/NO2/average/'
month_out = ['01', '02', '03', '04', '05', '06', '07', '08', '09', '10', '11', '12']
season_out = ['spring', 'summer', 'autumn', 'winter']
geo_info = [-180.0, 0.25, 0, 90.0, 0, 0.25]
year_min = 9999
year_max = 0
file_list = []
# 获取所有.he5文件,并统计年份
if not os.path.exists(output_directory):
os.mkdir(output_directory)
for root, dirs, files in os.walk(input_directory):
for file in files:
if file.endswith('.he5'):
file_list.append(os.path.join(root, file))
file_name = os.path.basename(os.path.join(root, file))
year_temp = int(file_name[19:23])
if year_temp < year_min:
year_min = year_temp
if year_temp > year_max:
year_max = year_temp
# print(year_min, year_max)
year_n = year_max - year_min + 1
# 月份存储数组初始ar_max化
data_total_month = np.full((720, 1440, 12), 0.0)
data_valid_month = np.full((720, 1440, 12), 0.0)
# 季节存储数组初始化
data_total_season = np.full((720, 1440, 4), 0.0)
data_valid_season = np.full((720, 1440, 4), 0.0)
season_pos = np.array([3, 3, 0, 0, 0, 1, 1, 1, 2, 2, 2, 3])
# 年份存储数组初始化
data_total_year = np.full((720, 1440, year_n), 0.0)
data_valid_year = np.full((720, 1440, year_n), 0.0)
for file_i in file_list:
current_file = h5.File(file_i, "r")
data_temp = np.array(current_file['/HDFEOS/GRIDS/ColumnAmountNO2/Data Fields/ColumnAmountNO2TropCloudScreened'])
# print(data_temp.shape)
data_temp = ((data_temp > 0.0) * data_temp / ct.N_A) * (10.0 ** 10.0)
file_temp = os.path.basename(file_i)
layer_i = int(file_temp[24:26]) - 1
data_total_month[:, :, layer_i] += data_temp
data_temp_valid = (data_temp > 0.0).astype(float)
data_valid_month[:, :, layer_i] += data_temp_valid
season_i = season_pos[layer_i]
data_total_season[:, :, season_i] += data_temp
data_valid_season[:, :, season_i] += data_temp_valid
year_i = int(file_temp[19:23]) - year_min
data_total_year[:, :, year_i] += data_temp
data_valid_year[:, :, year_i] += data_temp_valid
data_valid_month = (data_valid_month > 0.0) * data_valid_month + (data_valid_month == 0.0) * (1.0)
data_avr_month = data_total_month / data_valid_month
data_valid_season = (data_valid_season > 0.0) * data_valid_season + (data_valid_season == 0.0) * (1.0)
data_avr_season = data_total_season / data_valid_season
data_valid_year = (data_valid_year > 0.0) * data_valid_year + (data_valid_year == 0.0) * (1.0)
data_avr_year = data_total_year / data_valid_year
for month_i in range(12):
out_name = output_directory + 'month_avr_' + month_out[month_i] + '.tiff'
driver = gdal.GetDriverByName('GTiff')
out_array = np.flipud(data_avr_month[:, :, month_i])
out_file = driver.Create(out_name, 1440, 720, 1, gdal.GDT_Float32)
out_file.GetRasterBand(1).WriteArray(out_array)
out_file.SetGeoTransform(geo_info)
srs = osr.SpatialReference()
srs.ImportFromEPSG(4326)
out_file.SetProjection(srs.ExportToWkt())
for season_i in range(4):
out_name = output_directory + 'season_avr_' + season_out[season_i] + '.tiff'
driver = gdal.GetDriverByName('GTiff')
out_array = np.flipud(data_avr_season[:, :, season_i])
out_file = driver.Create(out_name, 1440, 720, 1, gdal.GDT_Float32)
out_file.GetRasterBand(1).WriteArray(out_array)
out_file.SetGeoTransform(geo_info)
srs = osr.SpatialReference()
srs.ImportFromEPSG(4326)
out_file.SetProjection(srs.ExportToWkt())
for year_i in range(year_n):
out_name = output_directory + 'year_avr_' + str(year_min + year_i) + '.tiff'
driver = gdal.GetDriverByName('GTiff')
out_array = np.flipud(data_avr_year[:, :, year_i])
out_file = driver.Create(out_name, 1440, 720, 1, gdal.GDT_Float32)
out_file.GetRasterBand(1).WriteArray(out_array)
out_file.SetGeoTransform(geo_info)
srs = osr.SpatialReference()
srs.ImportFromEPSG(4326)
out_file.SetProjection(srs.ExportToWkt())
if __name__ == '__main__':
main()
至于Python代码时间有限我就不进行对比了,我的python代码与我的IDL代码的图像结果几乎完全一致。(当然,思路也是基本相似;而实验给定的python代码应该也与实验给定IDL代码实现思路类似我没有细看,希望大家给出指正)