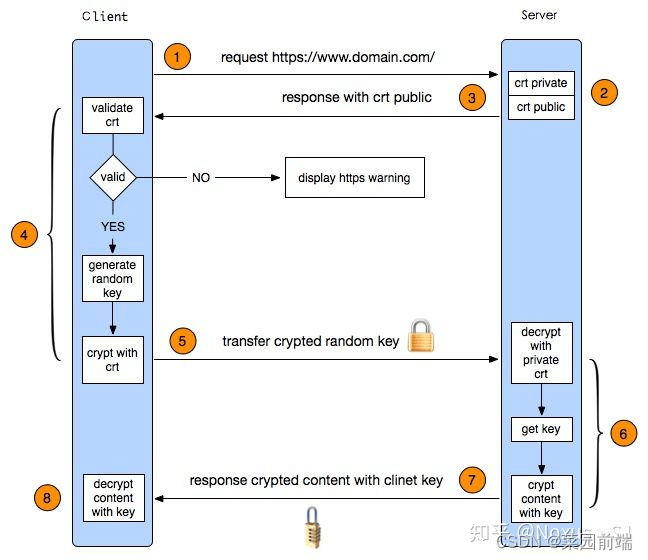
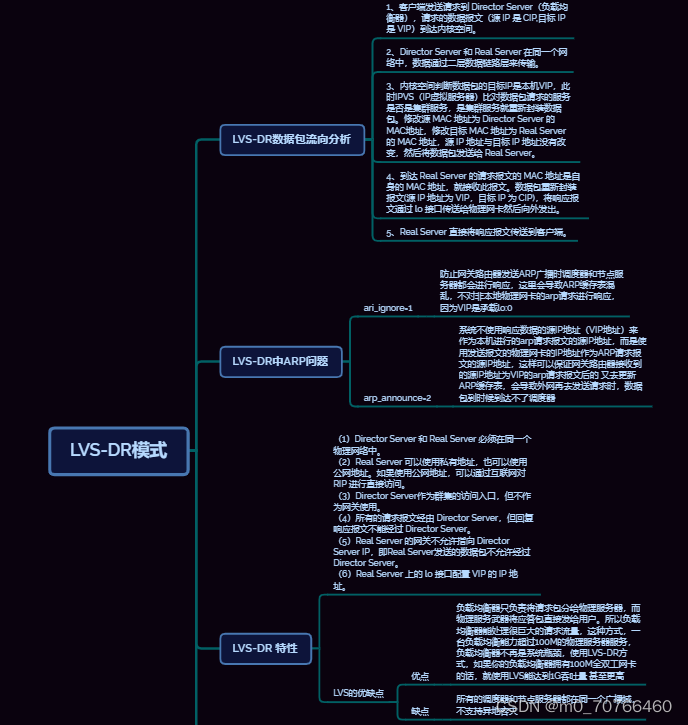
代码来自闵老师”日撸 Java 三百行(61-70天)
日撸 Java 三百行(61-70天,决策树与集成学习)_闵帆的博客-CSDN博客
本次代码的实现是基于高斯密度,ALEC算法原文是基于密度峰值,同样是基于密度聚类,稍微还是有一些差别。
基于聚类的主动学习的基本思想如下:
Step 1. 将对象按代表性递减排序;
Step 2. 假设当前数据块有 N个对象, 选择最具代表性的前个查询其标签 (类别).
Step 3. 如果这个标签具有相同类别, 就认为该块为纯的, 其它对象均分类为同一类别. 结束.
Step 4. 如果不纯,将当前块划分为两个子块, 分别 Goto Step 3.
package machinelearning.activelearning;
import java.io.FileReader;
import java.util.Arrays;
import weka.core.Instances;
public class Alec {
/**
* The whole data set.
*/
Instances dataset;
/**
* The maximal number of queries that can be provided.
*/
int maxNumQuery;
/**
* The actual number of queries.
*/
int numQuery;
/**
* The radius, also dc in the paper. It is employed for density computation.
*/
double radius;
/**
* The densities of instances, also rho in the paper.
*/
double[] densities;
/**
* Distance to master
*/
double[] distanceToMaster;
/**
* Sorted indices, where the first element indicates the instance with the biggest density.
*/
int[] descendantDensities;
/**
* Priority
*/
double[] priority;
/**
* The maximal distance between any pair of points.
*/
double maximalDistance;
/**
* Who is my master?
*/
int[] masters;
/**
* Predicted labels.
*/
int[] predictedLabels;
/**
* Instance status. 0 for unprocessed, 1 for queried, 2 for classified.
*/
int[] instanceStatusArray;
/**
* The descendant indices to show the representativeness of instances in a descendant order.
*/
int[] descendantRepresentatives;
/**
* Indicate the cluster of each instance. It is only used in clusterInTwo(int[]);
*/
int[] clusterIndices;
/**
* Blocks with size no more than this threshold should not be split further.
*/
int smallBlockThreshold = 3;
/**
* *********************************************************
* The constructor.
* @param paraFilename
* *********************************************************
*/
public Alec(String paraFilename) {
// TODO Auto-generated constructor stub
try {
FileReader tempReader = new FileReader(paraFilename);
dataset = new Instances(tempReader);
dataset.setClassIndex(dataset.numAttributes() - 1);
tempReader.close();
} catch (Exception e) {
// TODO: handle exception
System.out.println(e);
System.exit(0);
}//of try
computeMaximalDistance();
clusterIndices = new int[dataset.numInstances()];
}//of the constructor
/**
* ***********************************************************
* * Merge sort in descendant order to obtain an index array. The original
* array is unchanged. The method should be tested further. <br>
* Examples: input [1.2, 2.3, 0.4, 0.5], output [1, 0, 3, 2]. <br>
* input [3.1, 5.2, 6.3, 2.1, 4.4], output [2, 1, 4, 0, 3].
*
* @param paraArray The original array
* @return The sorted indices.
* ***********************************************************
*/
public static int[] mergeSortToIndices(double[] paraArray) {
int tempLength = paraArray.length;
int[][] resultMatrix = new int[2][tempLength];
//Initialize(这里初始化第一组就够了,第二组的数据在排序的时候是从第一组复制的)
int tempIndex = 0;
for (int i = 0; i < tempLength; i++) {
resultMatrix[tempIndex][i] = i;
}//of for i
// Merge
int tempCurrentLength = 1;
// The indices for current merged groups.
int tempFirstStart, tempSecondStart, tempSecondEnd;
while (tempCurrentLength < tempLength) {
// Divide into a number of groups.
// Here the boundary is adaptive to array length not equal to 2^k.
//Math.ceil()的作用是上取整,返回的是总共分了多少个“两组”数据。每进行一次for循环,完成一轮归并,当while条件不满足时,已经完成排序。
//每一次是进行两组数的归并,所以每一轮的组数是(tempLength + 0.0)/(tempCurrentLength * 2);每一组数据的长度是tempCurrentLength
for (int i = 0; i < Math.ceil((tempLength + 0.0) / tempCurrentLength /2); i++) {
// Boundaries of the group
tempFirstStart = i * tempCurrentLength * 2;
tempSecondStart = tempFirstStart + tempCurrentLength;
tempSecondEnd = tempSecondStart + tempCurrentLength - 1;
if (tempSecondEnd >= tempLength) {
tempSecondEnd = tempLength - 1;
}//of if
// Merge this group
int tempFirstIndex = tempFirstStart;
int tempSecondIndex = tempSecondStart;
int tempCurrentIndex = tempFirstStart;
if (tempSecondStart >= tempLength) {
for (int j = tempFirstIndex; j < tempLength; j++) {
resultMatrix[(tempIndex + 1) % 2][tempCurrentIndex] = resultMatrix[tempIndex % 2][j];
tempFirstIndex ++;
tempCurrentIndex ++;
}//of for j
break;
}//of if
while ((tempFirstIndex <= tempSecondStart - 1) && (tempSecondIndex <= tempSecondEnd)) {
if (paraArray[resultMatrix[tempIndex % 2][tempFirstIndex]] <= paraArray[resultMatrix[tempIndex % 2][tempSecondIndex]]) {
resultMatrix[(tempIndex + 1) % 2][tempCurrentIndex] = resultMatrix[tempIndex % 2][tempSecondIndex];
tempSecondIndex ++;
}else {
resultMatrix[(tempIndex + 1) % 2][tempCurrentIndex] = resultMatrix[tempIndex % 2][tempFirstIndex];
tempFirstIndex ++;
}//of if
tempCurrentIndex ++;
}//of while
// Remaining part
for (int j = tempFirstIndex; j < tempSecondStart; j++) {
resultMatrix[(tempIndex + 1) % 2][tempCurrentIndex] = resultMatrix[tempIndex % 2][j];
tempCurrentIndex++;
}//of for j
for (int j = tempSecondIndex; j <= tempSecondEnd; j++) {
resultMatrix[(tempIndex + 1) % 2][tempCurrentIndex] = resultMatrix[tempIndex % 2][j];
tempCurrentIndex++;
}//of for j
}//of for i
tempCurrentLength *= 2;
tempIndex ++; //交替使用两个数组为当前组,另一个数组用于新一轮排序时复制索引号。
}//of while
return resultMatrix[tempIndex % 2];
}//of mergeSortToIndices
/**
* **********************************************************************
* The Euclidean distance between two instances. Other distance measures
* unsupported for simplicity.
*
* @param paraI The index of the first instance.
* @param paraJ The index of the second instance.
* @return The distance.
* **********************************************************************
*/
public double distance(int paraI, int paraJ) {
double resultDistance = 0;
double tempDifference;
for (int i = 0; i < dataset.numAttributes() - 1; i++) {
tempDifference = dataset.instance(paraI).value(i) - dataset.instance(paraJ).value(i);
resultDistance += tempDifference * tempDifference;
}//of for i
resultDistance = Math.sqrt(resultDistance);
return resultDistance;
}//of distance
/**
* *****************************************************************
* Compute the maximal distance. The result is stored in a member variable.
* *****************************************************************
*/
public void computeMaximalDistance() {
maximalDistance = 0;
double tempDistance;
for (int i = 0; i < dataset.numInstances(); i++) {
for (int j = 0; j < dataset.numInstances(); j++) {
tempDistance = distance(i, j);
if (maximalDistance < tempDistance) {
maximalDistance = tempDistance;
}//of if
}//of for j
}//of for i
System.out.println("maximalDistance = " + maximalDistance);
}//of computeMaximalDistance
/**
* ****************************************************************
* Compute the densities using Gaussian kernel.
* 这里不同于原文的密度峰值聚类,这里用的高斯核函数计算实例的密度。密度峰值是设定一个距离阈值,在距离范围内的实例个数就是当前实例的密度。
*
* @param paraBlock The given block.
* ****************************************************************
*/
public void computeDensitiesGaussian() {
System.out.println("radius = " + radius);
densities = new double[dataset.numInstances()];
double tempDistance;
for (int i = 0; i < dataset.numInstances(); i++) {
for (int j = 0; j < dataset.numInstances(); j++) {
tempDistance = distance(i, j);
densities[i] += Math.exp(-tempDistance * tempDistance /(radius * radius));
}//of for j
}//of for i
System.out.println("The densities are " + Arrays.toString(densities) + "\r\n");
}//of computeDensitiesGaussian
/**
* ***************************************************************
* Compute distanceToMaster, the distance to its master.
* ***************************************************************
*/
public void computeDistanceToMaster() {
distanceToMaster = new double[dataset.numInstances()];
masters = new int[dataset.numInstances()];
descendantDensities = new int[dataset.numInstances()];
instanceStatusArray = new int[dataset.numInstances()];
descendantDensities = mergeSortToIndices(densities);
distanceToMaster[descendantDensities[0]] = maximalDistance;
double tempDistance;
for (int i = 1; i < dataset.numInstances(); i++) {
// Initialize.
distanceToMaster[descendantDensities[i]] = maximalDistance;
//只有密度比自己大的才可能是自己的Master,所以排序在i以后实例不用计算。
for (int j = 0; j <= i - 1; j++) {
tempDistance = distance(descendantDensities[i], descendantDensities[j]);
if (distanceToMaster[descendantDensities[i]] > tempDistance) {
distanceToMaster[descendantDensities[i]] = tempDistance;
masters[descendantDensities[i]] = descendantDensities[j];
}//of if
}//of for j
}//of for i
System.out.println("First compute, masters = " + Arrays.toString(masters));
System.out.println("descendantDensities = " + Arrays.toString(descendantDensities));
}//of computeDistanceToMaster
/**
* ****************************************************************
* Compute priority. Element with higher priority is more likely to be
* selected as a cluster center. Now it is rho * distanceToMaster. It can
* also be rho^alpha * distanceToMaster.
* ****************************************************************
*/
public void computePriority() {
priority = new double[dataset.numInstances()];
for (int i = 0; i < dataset.numInstances(); i++) {
priority[i] = densities[i] * distanceToMaster[i];
}//of for i
}//of computePriority
/**
* *******************************************************************
* The block of a node should be same as its master. This recursive method is efficient.
* @param paraIndex The index of the given node.
* @return The cluster index of the current node.
* *******************************************************************
*/
public int coincideWithMaster(int paraIndex) {
if (clusterIndices[paraIndex] == -1) {
int tempMaster = masters[paraIndex];
clusterIndices[paraIndex] = coincideWithMaster(tempMaster);
}//of if
return clusterIndices[paraIndex];
}//of coincideWithMaster
/**
* *********************************************************************
* Cluster a block in two. According to the master tree.
*
* @param paraBlock The given block.
* @return The new blocks where the two most represent instances serve as the root.
* *********************************************************************
*/
public int[][] clusterInTwo(int[] paraBlock) {
// Reinitialize. In fact, only instances in the given block is considered.
Arrays.fill(clusterIndices, -1);
// Initialize the cluster number of the two roots.
//这里把数组paraBlock的前两个元素存储的序号,所对应的簇标签分别设置为0和1.
//paraBlock的前两个序号对应的原始数据集的实例标签如果相同,这里有没有影响???
for (int i = 0; i < 2; i++) {
clusterIndices[paraBlock[i]] = i;
}//of for i
for (int i = 0; i < paraBlock.length; i++) {
if (clusterIndices[paraBlock[i]] != -1) {
continue;
}//of if
//i实例和自己的Master具有相同的类标签。
clusterIndices[paraBlock[i]] = coincideWithMaster(masters[paraBlock[i]]);
}//of for i
//The sub blocks.
int[][] resultBlock = new int[2][];
int tempFirstBlockCount = 0;
//长度是clusterIndices.length,此时没在paraBlock中的实例,上面代码已经填充了-1.
//只有paraBlock[i]才有数据。如果这里长度设置为paraBlock.length,下面判断条件应该是clusterIndices[paraBlock[i]] == 0。
//否则i无法将所有实例全部遍历。
for (int i = 0; i <clusterIndices.length; i++) {
if (clusterIndices[i] == 0) {
tempFirstBlockCount++;
}//of if
}//of for i
resultBlock[0] = new int[tempFirstBlockCount];
resultBlock[1] = new int[paraBlock.length - tempFirstBlockCount];
// Copy. You can design shorter code when the number of clusters is greater than 2.
int tempFirstIndex = 0;
int tempSecondIndex = 0;
for (int i = 0; i < paraBlock.length; i++) {
if (clusterIndices[paraBlock[i]] == 0) {
resultBlock[0][tempFirstIndex] = paraBlock[i];
tempFirstIndex ++;
} else {
resultBlock[1][tempSecondIndex] = paraBlock[i];
tempSecondIndex ++;
}//of if
}//of for i
System.out.println("Split (" + paraBlock.length + ") instances "
+ Arrays.toString(paraBlock) + "\r\nto (" + resultBlock[0].length + ") instances "
+ Arrays.toString(resultBlock[0]) + "\r\nand (" + resultBlock[1].length
+ ") instances " + Arrays.toString(resultBlock[1]));
return resultBlock;
}//of clusterInTwo
/**
* **************************************************************
* Classify instances in the block by simple voting.
*
* @param paraBlock The given block.
* **************************************************************
*/
public void vote(int[] paraBlock) {
int[] tempClassCounts = new int[dataset.numClasses()];
for (int i = 0; i < paraBlock.length; i++) {
if (instanceStatusArray[paraBlock[i]] == 1) {
//"1"代表可查询标签的实例。
tempClassCounts[(int)dataset.instance(paraBlock[i]).classValue()]++;
}//of if
}//of for i
int tempMaxClass = -1;
int tempMaxCount = -1;
for (int i = 0; i < tempClassCounts.length; i++) {
if (tempMaxCount < tempClassCounts[i]) {
tempMaxCount = tempClassCounts[i];
tempMaxClass = i;
}//of if
}//of for i
// Classify unprocessed instances.
for (int i = 0; i < paraBlock.length; i++) {
if (instanceStatusArray[paraBlock[i]] == 0) {
predictedLabels[paraBlock[i]] = tempMaxClass;
instanceStatusArray[paraBlock[i]] = 2;
}//of if
}//of for i
}//of vote
/**
* *****************************************************************************************************
* Cluster based active learning. Prepare for
*
* @param paraRatio The ratio of the maximal distance as the dc.
* @param paraMaxNumQuery The maximal number of queries for the whole dataset.
* @param paraSmallBlockThreshold The small block threshold.
* *****************************************************************************************************
*/
public void clusterBasedActiveLearning(double paraRatio, int paraMaxNumQuery, int paraSmallBlockThreshold) {
radius = maximalDistance * paraRatio;
smallBlockThreshold = paraSmallBlockThreshold;
maxNumQuery = paraMaxNumQuery;
predictedLabels = new int[dataset.numInstances()];
for (int i = 0; i < dataset.numInstances(); i++) {
predictedLabels[i] = -1;
}//of for i
computeDensitiesGaussian();
computeDistanceToMaster();
computePriority();
descendantRepresentatives = mergeSortToIndices(priority);
System.out.println("descendantRepresentatives = " + Arrays.toString(descendantRepresentatives));
numQuery = 0;
clusterBasedActiveLearning(descendantRepresentatives);
}//of clusterBasedActiveLearning
/**
* *******************************************************************************************************************
* Cluster based active learning.
*
* @param paraBlock The given block. This block must be sorted according to the priority in descendant order.
* *******************************************************************************************************************
*/
public void clusterBasedActiveLearning(int[] paraBlock) {
System.out.println("clusterBasedActiveLearning for block " + Arrays.toString(paraBlock));
// Step 1. How many labels are queried for this block.
int tempExpectedQueries = (int)Math.sqrt(paraBlock.length);
int tempNumQuery = 0;
for (int i = 0; i < paraBlock.length; i++) {
if (instanceStatusArray[paraBlock[i]] == 1) {
tempNumQuery ++;
}//of if
}//of for i
// Step 2. Vote for small blocks.
if ((tempNumQuery >= tempExpectedQueries) && (paraBlock.length <= smallBlockThreshold)) {
System.out.println("" + tempNumQuery + " instances are queried, vote for block: \r\n"
+ Arrays.toString(paraBlock));
vote(paraBlock);
return;
}//of if
// Step 3. Query enough labels.
for (int i = 0; i < tempExpectedQueries; i++) {
if (numQuery >= maxNumQuery) {
System.out.println("No more queries are provided, numQuery = " + numQuery + ".");
vote(paraBlock);
return;
}//of if
if (instanceStatusArray[paraBlock[i]] == 0) {
instanceStatusArray[paraBlock[i]] = 1;
predictedLabels[paraBlock[i]] = (int)dataset.instance(paraBlock[i]).classValue();
numQuery ++;
}//of if
}//of for i
//Step 4. Pure?
int tempFirstLabel = predictedLabels[paraBlock[0]];
boolean tempPure = true;
for (int i = 1; i < tempExpectedQueries; i++) {
if (predictedLabels[paraBlock[i]] != tempFirstLabel) {
tempPure = false;
break;
}//of if
}//of for i
if (tempPure) {
System.out.println("Classify for pure block: " + Arrays.toString(paraBlock));
for (int i = tempExpectedQueries; i < paraBlock.length; i++) {
if (instanceStatusArray[paraBlock[i]] == 0) {
instanceStatusArray[paraBlock[i]] = 2;
predictedLabels[paraBlock[i]] = tempFirstLabel;
}//of if
}//of for i
return;
}//of if
// Step 5. Split in two and process them independently.
int[][] tempBlocks = clusterInTwo(paraBlock);
for (int i = 0; i < 2; i++) {
clusterBasedActiveLearning(tempBlocks[i]);
}//of for i
}//of clusterBasedActiveLearning
/**
******************************************************
* Show the statistics information.
******************************************************
*/
public String toString() {
int[] tempStatusCounts = new int[3];
double tempCorrect = 0;
for (int i = 0; i < dataset.numInstances(); i++) {
tempStatusCounts[instanceStatusArray[i]]++;
if (predictedLabels[i] == (int) dataset.instance(i).classValue()) {
tempCorrect ++;
}//of if
}//of for i
String resultString = "(unhandled, queried, classified) = " + Arrays.toString(tempStatusCounts);
resultString += "\r\nCorrect = " + tempCorrect + ", accuracy = " + (tempCorrect / dataset.numInstances());
return resultString;
}//ofr toString
/**
* ********************************************************************
* The entrance of the program.
*
* @param args
* ********************************************************************
*/
public static void main(String args[]) {
long tempStart = System.currentTimeMillis();
System.out.println("Starting ALEC.");
String arffFileName = "E:/Datasets/UCIdatasets/其他数据集/iris.arff";
Alec tempAlec = new Alec(arffFileName);
// The settings for iris
tempAlec.clusterBasedActiveLearning(0.15, 30, 3);
System.out.println(tempAlec);
long tempEnd = System.currentTimeMillis();
System.out.println("Runtime: " + (tempEnd - tempStart) + "ms.");
}//of main
}//of Alec
部分对代码的理解已经标注在备注里。