欢迎关注我的CSDN:https://spike.blog.csdn.net/
本文地址:https://blog.csdn.net/caroline_wendy/article/details/131201456
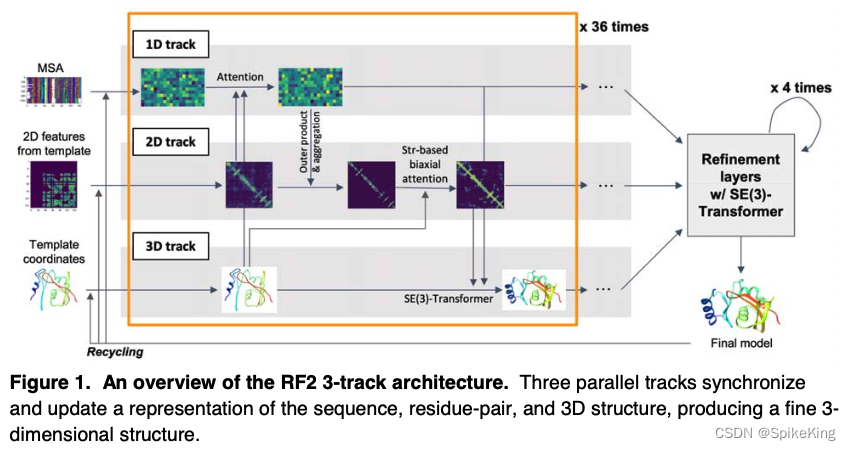
RoseTTAFold2 是蛋白质结构预测算法,利用了深度学习和三维几何建模的技术,能够快速准确地预测蛋白质的三级结构。RoseTTAFold2 是在 RoseTTAFold 的基础上改进的,增加了对多肽段的处理,提高了对复杂蛋白质的预测能力,同时也降低了计算资源的需求。
Paper: Efficient and accurate prediction of protein structure using RoseTTAFold2,2023.5.25
预测结果的数据路径:output/rf2-monomer/,56个样本
以T1104-D1_A117为例,具体格式如下:
hhblits/
log/
models/
T1104-D1_A117_1.atab*
T1104-D1_A117_1.fa*
T1104-D1_A117_1.hhr*
T1104-D1_A117_1.msa0.a3m*
PDB模型,位于models,PDB数据文件名,即model_00_pred.pdb。
model_00.json
model_00.npz
model_00_pred.pdb
结果导出脚本:main_rf2_results_parser.py
python scripts/main_rf2_results_parser.py -i output/rf2-monomer/ -o mydata/results_v2/casp15-fasta-56-outputs-rf2-results
源码如下:
#!/usr/bin/env python
# -- coding: utf-8 --
"""
Copyright (c) 2022. All rights reserved.
Created by C. L. Wang on 2023/6/14
"""
import argparse
import collections
import json
import os
import shutil
import sys
from pathlib import Path
from tqdm import tqdm
p = os.path.dirname(os.path.dirname(os.path.abspath(__file__)))
if p not in sys.path:
sys.path.append(p)
from protein_utils.pdb_utils import get_plddt_from_pdb
from myutils.project_utils import traverse_dir_files, mkdir_if_not_exist
class MainRf2ResultsParser(object):
"""
RoseTTAFold2 的结果处理,以利于后续评估
"""
def __init__(self):
pass
def process(self, input_dir, output_dir):
print(f"[Info] 输入文件夹: {input_dir}")
print(f"[Info] 输出文件夹: {output_dir}")
mkdir_if_not_exist(output_dir)
path_list = traverse_dir_files(input_dir, ext="pdb")
print(f"[Info] 样本数量: {len(path_list)}")
fasta_dict = collections.defaultdict(list)
for path in tqdm(path_list, desc="[Info] PDB"):
tmp_items = path.split("/")
fasta_name = tmp_items[-3]
fasta_dict[fasta_name].append(path)
print(f"[Info] FASTA数量: {len(fasta_dict.keys())}")
for fasta_name in tqdm(fasta_dict.keys(), desc="[Info] FASTA"):
fasta_path_list = fasta_dict[fasta_name]
# 构建文件夹
out_fasta_dir = os.path.join(output_dir, fasta_name)
mkdir_if_not_exist(out_fasta_dir)
pdb_score_list = []
for path in fasta_path_list:
file_name = os.path.basename(path)
# 复制文件
shutil.copy(path, os.path.join(out_fasta_dir, file_name))
# 写入文件
plddt_val = float(get_plddt_from_pdb(path))
pdb_name = file_name.split(".")[0]
pdb_score_list.append((pdb_name, plddt_val))
# 写入评估结果的JSON文件
pdb_score_list = sorted(pdb_score_list, key=lambda x: x[1], reverse=True)
score_map_path = os.path.join(out_fasta_dir, "ranking_debug.json")
with open(score_map_path, 'w') as f:
score_map_dict = {
"plddts": dict(pdb_score_list),
"order": [x[0] for x in pdb_score_list],
}
json.dump(score_map_dict, f, indent=4, sort_keys=True)
def main():
parser = argparse.ArgumentParser()
parser.add_argument(
"-i",
"--input_dir",
help="the dir from rosettafold2 output.",
type=Path,
required=True,
)
parser.add_argument(
"-o",
"--output-dir",
help="output dir path.",
type=Path,
required=True
)
args = parser.parse_args()
input_dir = str(args.input_dir)
output_dir = str(args.output_dir)
mkdir_if_not_exist(output_dir)
assert os.path.isdir(input_dir) and os.path.isdir(output_dir)
rrp = MainRf2ResultsParser()
rrp.process(input_dir, output_dir)
if __name__ == '__main__':
main()